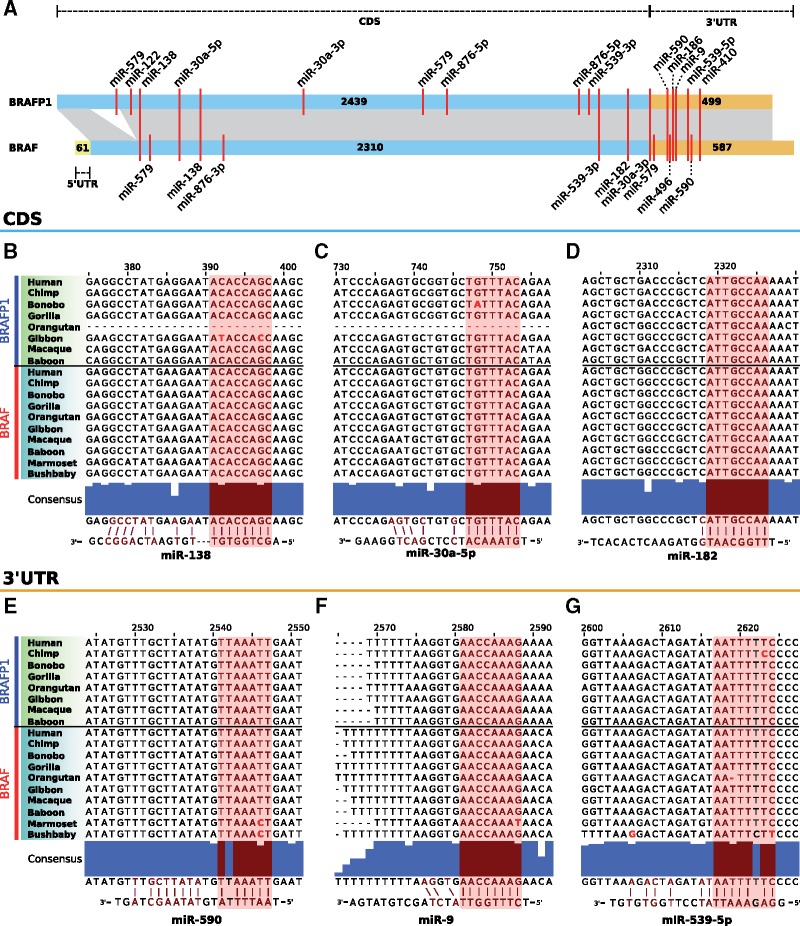
Fig. 2.
Predicted MRE locations and sequence conservation in BRAF and BRAFP1. (A) Locations of MREs for miRNAs confirmed and/or predicted to regulate BRAF and BRAFP1 are shown with respect to the gene and pseudogene structures. MiRanda target prediction (Enright et al. 2003) was used to locate the putative MREs. Red bars spanning both elements indicate MREs present in both, whereas red bars on one element only indicate element-specific MREs. The full list of miRNAs and number of species that each MRE is predicted to be present in can be found in supplementary table 1, Supplementary Material online. Lengths of each region, corresponding to the human sequence, are indicated. Predicted conserved MREs across BRAF and BRAFP1 CDS (B–D) and 3′UTR (E–G) in Catarrhini primates. Red shading indicates the seed region, and blue consensus bars for each base denote the proportion of sequences that match the consensus sequence. The position along sequences relative to human BRAFP1 is indicated. The miRNA mature sequences are also shown directly below the consensus sequences. Bases in bold red font indicate substitutions predicted to be detrimental to miRNA binding. The long deletion in the pseudo-CDS of orangutan is supported by BLAST searches of two independent genome builds and inspection of intact contigs. Alignments were visualized using JalView (Waterhouse et al. 2009).