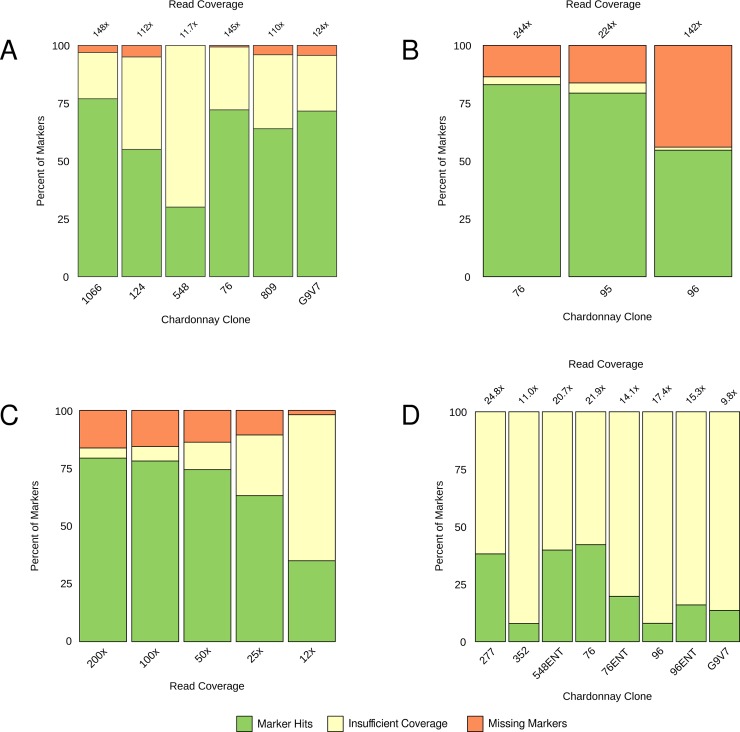
Fig 5. Validation of clonal markers for Chardonnay.
Results of marker detection pipeline run on four WGS sequencing datasets. The percentage breakdown of clonal markers is shown as either markers identified in the datasets (marker hits), missing from the datasets (missing markers), or missing with low coverage at marker loci (insufficient coverage). (A) Moderate coverage sequencing replicates of marker discovery clonal material. (B) High-coverage sequencing of independently-sourced clonal material. (C) Coverage subsampling of independently-sourced clonal material (clone 95 from Fig 5B). (D) Low-coverage sequencing of DNA derived from several independently-sourced clones (‘ENT’ denotes ENTAV-INRA source material).