Abstract
Objectives
KLHL6 is a recently described BTB-Kelch protein with selective expression in lymphoid tissues and is most strongly expressed in germinal center B cells.
Methods
Using gene expression profiling as well as immunohistochemistry with an anti-KLHL6 monoclonal antibody, we have characterized the expression of this molecule in normal and neoplastic tissues. Protein expression was evaluated in 1,058 hematopoietic neoplasms.
Results
Consistent with its discovery as a germinal center marker, KLHL6 was positive mainly in B-cell neoplasms of germinal center derivation, including 95% of follicular lymphomas (106/112). B-cell lymphomas of non–germinal center derivation were generally negative (0/33 chronic lymphocytic leukemias/small lymphocytic lymphomas, 3/49 marginal zone lymphomas, and 2/66 mantle cell lymphomas).
Conclusions
In addition to other germinal center markers, including BCL6, CD10, HGAL, and LMO2, KLHL6 immunohistochemistry may prove a useful adjunct in the diagnosis and future classification of B-cell lymphomas.
Keywords: Hematopathology, KLHL6, Immunohistochemistry, Lymphoma
Immunohistochemical staining for specific markers of differentiation has become a critical component in the classification and diagnosis of hematopoietic neoplasms. Markers of germinal center differentiation in B-cell neoplasms are of special interest, both because several B-cell lymphomas correspond to the germinal center phase of B-cell development and also because the germinal center phenotype appears to define a prognostically significant subset of diffuse large B-cell lymphomas (DLBCLs).1
The KLHL6 gene was first described in a complementary DNA subtraction screen to identify factors that might be involved in developmental immunoglobulin gene mutation in ovine B cells. Subsequently, specific overexpression of this gene was shown in human germinal centers.2 KLHL6 belongs to the BTB-Kelch family of proteins. Family members contain BTB domains (for Broad-Complex, Tramtrack, and Bric-a-brac) as well as Kelch motifs (named for their structural similarity to the kelch gene in Drosophila melanogaster). Both domains are protein-protein interaction domains. There is relatively little published literature concerning the function of these proteins, especially in humans, but they appear to be involved in a diverse set of cellular functions, particularly regulation of protein turnover, as some act as substrate recognition components for ubiquitin ligases.3,4 In mammals, a member of the BTB-Kelch family known as KEAP1 is required for Nrf2-dependent regulation of antioxidant and detoxification-related genes.5 Haploinsufficiency for KLHL10 causes infertility,6 and GAN (gigaxonin) mutations underlie the disorder giant axon neuropathy.7 KLHL24, the closest family member to KLHL6, regulates protein levels of cytokeratin 14, and dominant mutations in its gene contribute to epidermolysis bullosa.8
KLHL6 is highly conserved throughout vertebrates. In mammals, evidence suggests that KLHL6 functions in B-cell signaling and is required for the germinal center response. Although the gene is expressed throughout B-cell ontogeny, KLHL6–/– mice have apparently normal early B-cell development. However, mature B-cell numbers are reduced, and germinal center formation, antigen-dependent B-cell proliferation, and formation of memory B cells are all impaired.9 A more recent study identified KLHL6 protein as a component of the B-cell receptor signalosome.10
Levels of KLHL6 messenger RNA (mRNA) are markedly increased in tissues and purified cells enriched for germinal center B cells.2 Because mature B-cell lymphomas often recapitulate to some extent the phenotypes of mature B-cell subsets, and because the distinction between germinal center and non–germinal center B cells also appears to have significance within existing disease entities as currently defined,1 we sought to explore the expression of this molecule in a variety of B-cell lymphomas. In addition, it has been recently reported that recurrent somatic mutations, sometimes biallelic, in the KLHL6 gene are present in some B-cell neoplasms, including DLBCL,11 chronic lymphocytic leukemia,12,13 and marginal zone lymphoma,14 further highlighting its likely biologic significance.
Materials and Methods
Gene Expression Analysis
Data for RNA sequencing analysis of normal tissues were obtained from RNAseq Atlas.15 Raw cel files for publicly available Affymetrix (Santa Clara, CA) U133 plus 2.0 gene expression microarray data for normal immune cells (GSE49910),16 normal B cells and B-cell malignancies (GSE12453),17 and DLBCL tumors (GSE10846)18 were obtained from the gene expression omnibus. Data were robust multiarray average normalized using the ExpressionFileCreator module of GenePattern.19 Scores to categorize DLBCL tumors by cell of origin subtype were calculated according to the Wright algorithm.20 For Affymetrix U133B microarrays, only a single probe set is present for KLHL6. For Affymetrix U133 plus two microarrays, intensities from the four probe sets for KLHL6 (1555275_a_at, 1560396_at, 1560397_s_at, 228167_at) were averaged for use in survival analysis. Association between KLHL6 transcript level and survival was tested in two cyclophosphamide/doxorubicin/vincristine/prednisone (CHOP)-treated and two R-CHOP (CHOP plus rituximab)–treated cohorts using Cox regression. Survival associations were also tested in the Visco et al21 data set by dichotomizing samples into those with expression values higher or lower than the data set median and using a log-rank test. The same grouping and log-rank test was used for independent analysis of activated B-cell (ABC)–like and germinal center B-cell (GCB)–like subtypes, according to the classification reported in their study, which had been shown to be valid in a direct comparison of fresh/frozen and formalin-fixed, paraffin-embedded (FFPE)–derived RNA.22
Tissue Samples
FFPE tissue samples were obtained from the archives of the Department of Pathology, Stanford University School of Medicine, Stanford, California. A total of 1,105 tumors, including 1,044 lymphoid neoplasms, were studied. Tissue microarrays (TMAs) incorporating FFPE tissue were constructed as previously described.23 All tissues were obtained prior to treatment, and institutional review board approval from Stanford University was obtained for this study. For expression in normal hematopoietic tissues, whole tissue sections of normal human spleen, thymus, bone marrow, and tonsil were used. For expression in nonhematopoietic tissues, a TMA containing samples of normal uterus, thyroid, testis, stomach, salivary gland, prostate, parathyroid, pancreas, lung, liver, kidney, heart, colon, brain, adrenal gland, skeletal muscle, and placenta was used. Hematolymphoid neoplasia was classified according to the 2016 World Health Organization classification.24
Monoclonal KLHL6 Antibody Validation
The 92C murine monoclonal antibody was generated using recombinant polyhistidine-tagged human KLHL6 protein as the antigen. The antibody was able to specifically identify overexpressed human V5-tagged KLHL6 protein in HEK293T cells by Western blotting (Supplemental Figure 1; all supplemental materials can be found at the American Journal of Clinical Pathology online) and immunohistochemistry on frozen cytospin preparations (Supplemental Figure 2). Although there are 42 KLHL family genes in humans, these are quite diverse. The two most closely related family members are KLHL24 and KLHL35, which show only 42.5% and 33.2% sequence identity at the protein level, respectively, indicating a very low likelihood of meaningful cross-reactivity.
Immunohistochemistry
The antibodies against KLHL6, HGAL, CD10, and BCL6 used here and the conditions used for immunohistochemistry are shown in Table 1. All immunostaining was performed on 4-µm-thick sections of FFPE tissues in the form of TMAs or as whole sections. Automated immunostainers were used: Leica BOND-MAX (Leica Biosystems, Buffalo Grove, IL) and Ventana BenchMark (Ventana Medical Systems, Tucson, AZ). Cases were considered positive for KLHL6 if at least 5% of tumor cells were stained and strong positive if there was strong, diffuse staining in greater than 30% of tumor cells. This cutoff was selected as we felt it would most reproducibly separate cases into three groups. The other stains were considered positive if more than 30% of cells stained. Cases were scored by two pathologists (C.A.K. and Y.N.); discrepant cases were jointly reviewed using a multiheaded microscope and a final score assigned.
Table 1.
Reagents and Methods for Immunohistochemistry
| Antibody/Clone | Dilution | Automated Staining Platform and Conditions | Source |
|---|---|---|---|
| KLHL6/92C | 1:3 | Leica Bond-Max; ER2 protocol |
Courtesy of Giovanna Roncador |
| HGAL/MRQ-49 | 1:100 | Leica Bond-Max; ER2 protocol |
Cell Marque, Rocklin, CA |
| CD10/56C6 | 1:20 | Leica Bond-Max; ER2 protocol |
Novocastra, Newcastle-upon-Tyne, UK |
| BCL6/GL191E/A8 | 1:100 | Ventana BenchMark; mild retrieval |
Cell Marque, Rocklin, CA |
Data Analysis and Visualization
Images of normal human hematolymphoid tissue and TMA immunohistochemical staining results were acquired using a Nikon Eclipse E1000 microscope (Nikon, Tokyo, Japan) equipped with ×4, ×10, ×20, ×40, and ×60 objective lenses with numerical apertures ranging from 0.05 to 0.90. Images were captured with a SPOT flex mosaic 15.2 digital camera and software (Diagnostic Instruments, Sterling Heights, MI).
Results
KLHL6 Gene Expression in Normal and Neoplastic Tissues
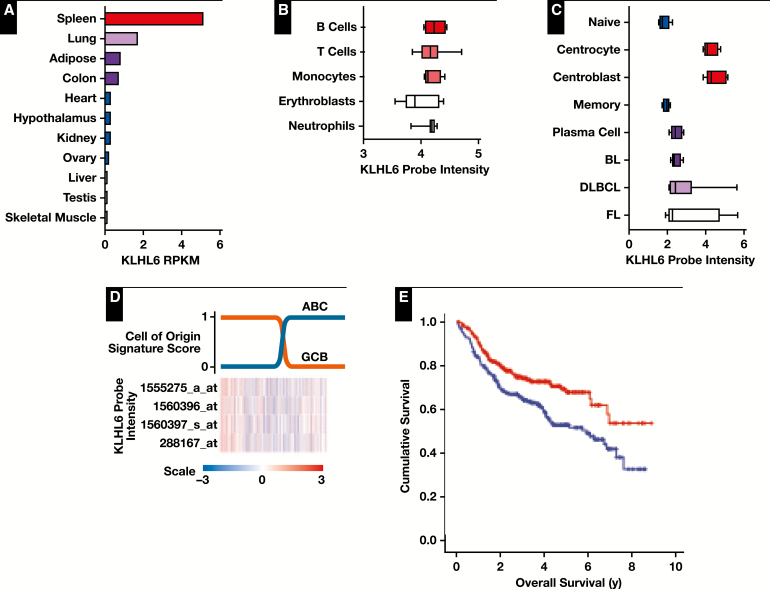
KLHL6 gene expression was highest in spleen among normal organs evaluated and was specifically upregulated in germinal center B cells, in agreement with previous reports2Figure 1A, Figure 1B, and Figure 1C. The data show that some expression of KLHL6 also occurs in non–B cells (Figure 1B), at least at the mRNA level, suggesting either that it has a broader role in other cell types at a lower expression level or that protein expression is suppressed at the posttranscriptional level. Among a small number of cases of DLBCL, follicular lymphoma, and Burkitt lymphoma (nine, five, and five cases, respectively), KLHL6 was expressed at similar levels, although a few cases of follicular lymphoma and DLBCL showed markedly increased levels (Figure 1C). In cases of DLBCL, RNA expression levels of KLHL6 did not differentiate strongly between the germinal center and activated B cell types Figure 1D. Despite this, however, expression levels were strongly prognostic, with higher expression levels predicting longer overall survival Figure 1E in the overall cohort as well as within both the GCB-like Figure 1F and ABC-like Figure 1G subtypes.
Figure 1.
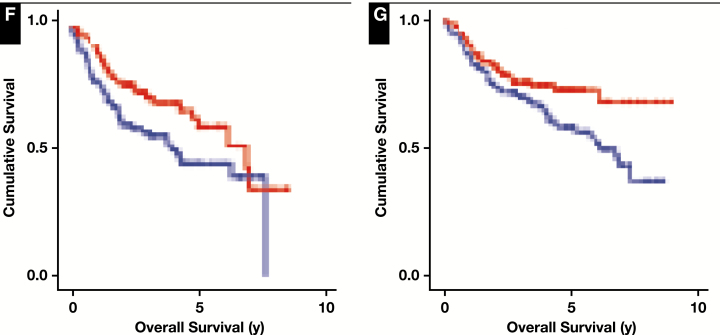
Transcript abundance of KLHL6. A, Measurement of KLHL6 (NM_130446) by RNAseq in normal human tissues shows strongest expression in the spleen. Data presented in reads per kilobase mapped (RPKM). Colors added to emphasize high values only. B, Gene expression microarray analysis of normal immune cells shows highest KLHL6 (1555275_a_at) expression in B cells. Data presented as log2-transformed probe intensity. Coloring of the lineages has no special significance. C, KLHL6 is highly expressed in germinal center B cells (centrocytes and centroblasts) compared with normal naive B cells, memory B cells, and plasma cells. Data presented as log2-transformed probe intensity. BL, Burkitt lymphoma; FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma. D, Despite being strongly expressed in normal germinal center B cells, KLHL6 is not strongly associated with the germinal center B-cell (GCB)–like subtype of DLBCL. Row-normalized probe intensities for KLHL6 are shown for 470 tumors from Visco et al21 ordered from left to right according to increasing probability of being an activated B-cell (ABC)–like subtype (blue) and decreasing probability of being a GCB-like subtype (orange). Classification scores are calculated based upon gene expression profiling data according to the Wright algorithm. The heatmap for KLHL6 expression is row normalized, with red indicating higher expression and blue indicating lower expression. E, Kaplan-Meier survival curves of 470 DLBCL tumors dichotomized in tumors having expression greater than (red) or less than (blue) the data set median. Higher expression was associated with superior overall survival (log-rank P = .002).
Figure 1 (cont) F, Kaplan-Meier survival curve of GCB-like tumors (n = 179) with expression greater than (red) or less than (blue) the data set median. Higher KLHL6 expression was able to predict superior overall survival within this subtype (log-rank P = .031). G, Kaplan-Meier survival curve of ABC-like tumors (n = 260) with expression greater than (red) or less than (blue) the data set median. Higher KLHL6 expression was able to predict superior overall survival within this subtype (log-rank P = .022).
KLHL6 Protein Expression in Normal Tissues
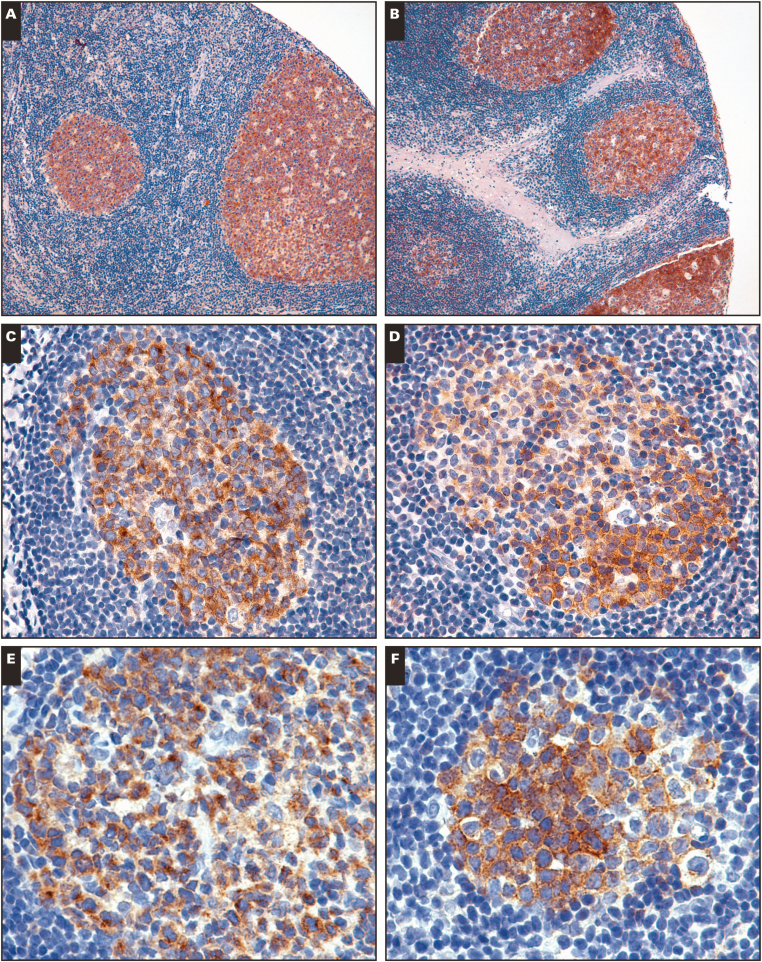
We used immunohistochemistry to determine the KLHL6 expression status of normal tissues. Sections of lymph nodes and tonsil showed cytoplasmic staining restricted to germinal centers, with variable Golgi accentuation Image 1A, Image 1C, and Image 1E. The staining pattern is similar to another cytoplasmic germinal center B-cell marker, HGAL Image 1B, Image 1D, and Image 1F. No significant staining was observed in thymus, spleen, bone marrow, or a wide variety of nonhematopoietic tissues (except when germinal centers were present).
Image 1.
KLHL6 reactivity in normal lymphoid tissues. A-C, KLHL6 immunostaining in normal tonsils demonstrating germinal center–restricted staining. D-F, HGAL shows a similar staining pattern in germinal centers. A, ×100. B, ×600. C, ×200. D, ×200. E, ×400. F, ×400.
KLHL6 Expression in Lymphomas
Immunohistochemistry for KLHL6 was performed on 667 non-Hodgkin B-cell lymphomas, 192 cases of classical and nodular lymphocyte-predominant Hodgkin lymphoma, and 246 other neoplasms, including plasma cell neoplasms, T-cell lymphomas, and nonhematopoietic tumors. Staining was observed in most follicular lymphomas as well as in a significant subset of DLBCLs and Hodgkin lymphomas; the data for all tumors tested are presented in Table 2.
Table 2.
KLHL6 Protein Expression by Immunohistochemistry in Human Neoplasms
| Neoplasm | No. of Cases | No. (%) Positive | No. (%) Strong Positive |
|---|---|---|---|
| Follicular lymphoma | 112 | 106 (94.6) | 76 (67.9) |
| Diffuse large B-cell lymphoma | 394 | 334 (84.8) | 182 (46.2) |
| Mantle cell lymphoma | 66 | 2 (3.0) | 2 (3.0) |
| Chronic lymphocytic lymphoma | 33 | 0 (0) | 0 (0) |
| Extranodal marginal zone lymphoma | 32 | 0 (0) | 0 (0) |
| Nodal marginal zone lymphoma | 9 | 3 (33.0) | 2 (22.0) |
| Splenic marginal zone lymphoma | 8 | 0 (0) | 0 (0) |
| Burkitt lymphoma | 6 | 4 (66.7) | 2 (33.3) |
| Lymphoplasmacytic lymphoma | 7 | 0 (0) | 0 (0) |
| Classical Hodgkin lymphoma | 181 | 72 (39.8) | 5 (2.8) |
| Nodular lymphocyte-predominant Hodgkin lymphoma | 11 | 11 (100.0) | 10 (50.0) |
| Plasma cell neoplasms | 139 | 10 (7.2) | 1 (0.7) |
| B lymphoblastic leukemia | 4 | 0 (0) | 0 (0) |
| T lymphoblastic leukemia | 9 | 0 (0) | 0 (0) |
| T-cell lymphomas | 30 | 1 (3.3) | 0 (0) |
| Acute myeloid leukemia | 11 | 0 (0) | 0 (0) |
| Extranodal natural killer/T-cell lymphomas | 3 | 0 (0) | 0 (0) |
| Langerhans cell histiocytosis | 3 | 1 (33.3) | 0 (0) |
| Nonhematopoietic neoplasms | 47 | 0 (0) | 0 (0) |
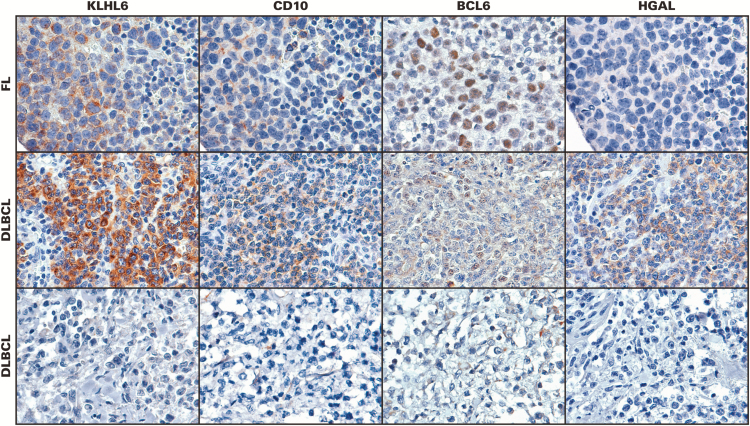
In follicular lymphoma, the KLHL6 antibody labeled tumor cells in follicles as well in interfollicular areas, paralleling the staining pattern seen with other well-characterized germinal center markers Image 2, top row). Cases of DLBCL showed wide variability of staining, from completely negative to very strong (Image 2, middle and bottom rows). B-cell lymphomas considered of non–germinal center origin were almost entirely negative, with rare exceptions Image 3A shows a KLHL6-positive case of mantle cell lymphoma). A subset of classical Hodgkin lymphomas also showed significant staining, with the antibody showing cytoplasmic reactivity in the large, atypical cells Image 3B, and similar staining was seen in nodular lymphocyte-predominant Hodgkin lymphoma Image 3C. Staining was generally weaker in these cases than in cases of follicular lymphoma or DLBCL. T-cell lymphomas were negative, although weak staining was observed in one case of transformed mycosis fungoides Image 3D.
Image 2.
KLHL6 immunohistochemistry with anti-KLHL6 monoclonal antibody in lymphomas. Top row: a case of follicular lymphoma stained with KLHL6, CD10, BCL6, and HGAL. Middle row: a case of diffuse large B-cell lymphoma (DLBCL) showing staining for KLHL6, CD10, BCL6, and HGAL. Bottom row: another case of DLBCL without expression of KLHL6, CD10, BCL6, or HGAL. (×200)
Image 3.
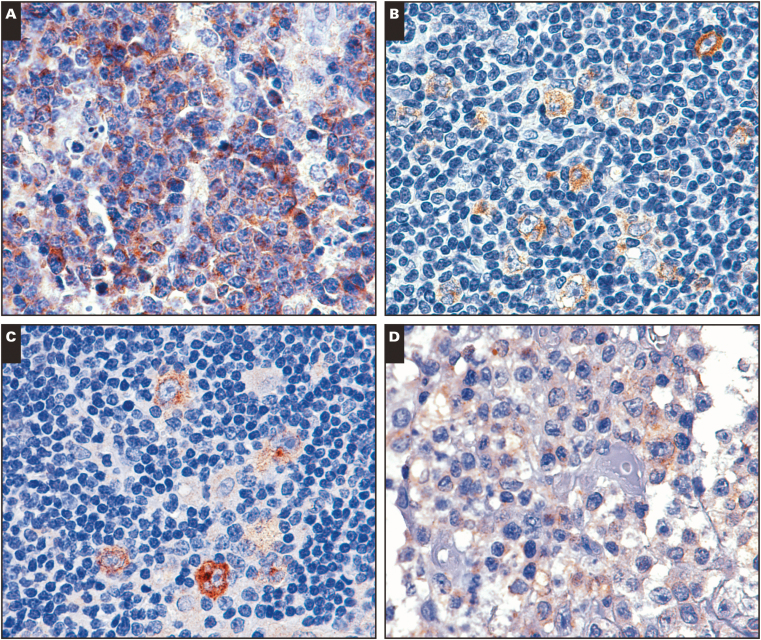
A, Unusual KLHL6 staining in a case of mantle cell lymphoma. B, KLHL6 staining in a case of classical Hodgkin lymphoma. C, KLHL6 staining in a case of nodular lymphocyte-predominant Hodgkin lymphoma. D, Weak KLHL6 staining in a case of transformed mycosis fungoides. (×200)
Expression of KLHL6 in Lymphomas Compared With Other Germinal Center Markers
Germinal center B-cell markers have diagnostic utility in the classification of small B-cell lymphomas. In addition, the expression of these markers in DLBCL is of particular interest, as a percentage of cases of de novo DLBCL appears to derive from germinal center B cells, and these cases (at least when defined by gene expression profiling) have a better prognosis independent of the usual clinicopathologic features.1 For these reasons, we compared the staining pattern of KLHL6 with three previously characterized markers now in routine clinical practice (CD10, BCL6, and HGAL).
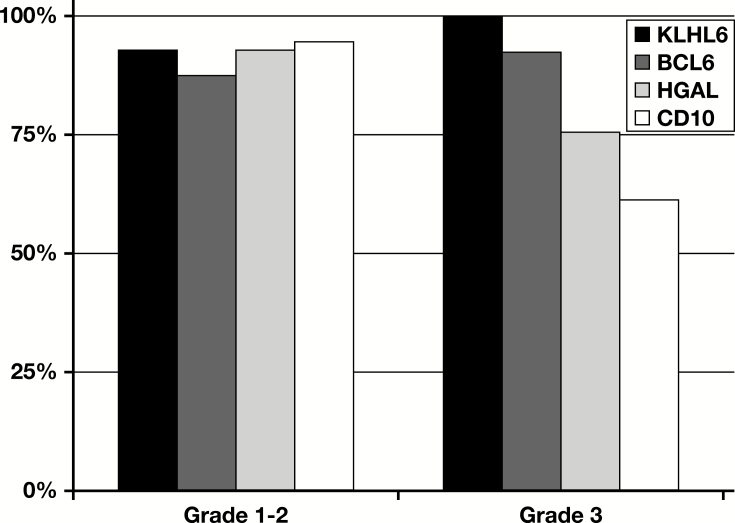
More than 75% of the 57 cases of low-grade (grades 1-2) follicular lymphoma that could be scored for all markers were positive for all four. However, in grade 3 follicular lymphoma, expression of one or more markers was lost in most cases, with loss of CD10 being the most frequent (38.5% of cases). In this sample of 38 cases of grade 3 follicular lymphoma, KLHL6 was positive in all cases Figure 2.
Figure 2.
Expression of germinal center markers in follicular lymphoma. Histogram showing the proportion of cases staining positive for each germinal center marker (CD10, BCL6, HGAL, and KLHL6) in grade 1 to 2 follicular lymphoma and in grade 3 follicular lymphoma.
In DLBCL, expression of each germinal center protein was seen in a subset of cases Table 3. In 57 cases of DLBCL, the expression of all four markers could be evaluated. The most common immunophenotypes (each seen in 10 cases or 15.7%) were absence of all four markers (Image 2, bottom row) or expression of all four markers (Image 2, middle row). The next most common pattern (nine cases or 15.8%) was expression of KLHL6, HGAL, and BCL6 without CD10, similar to the findings seen in high-grade follicular lymphoma Figure 3.
Table 3.
Immunohistochemistry for KLHL6 in 60 Cases of Diffuse Large B-Cell Lymphoma Compared With Other Germinal Center B-Cell Markers
| Score | KLHL6, No. | HGAL, No. | CD10, No. | BCL6, No. |
|---|---|---|---|---|
| Strong positive | 20 | 21 | 18 | 22 |
| Weak positive | 22 | 11 | 7 | 9 |
| Negative | 18 | 27 | 33 | 29 |
| Cannot assess | 0 | 1 | 2 | 0 |
Figure 3.
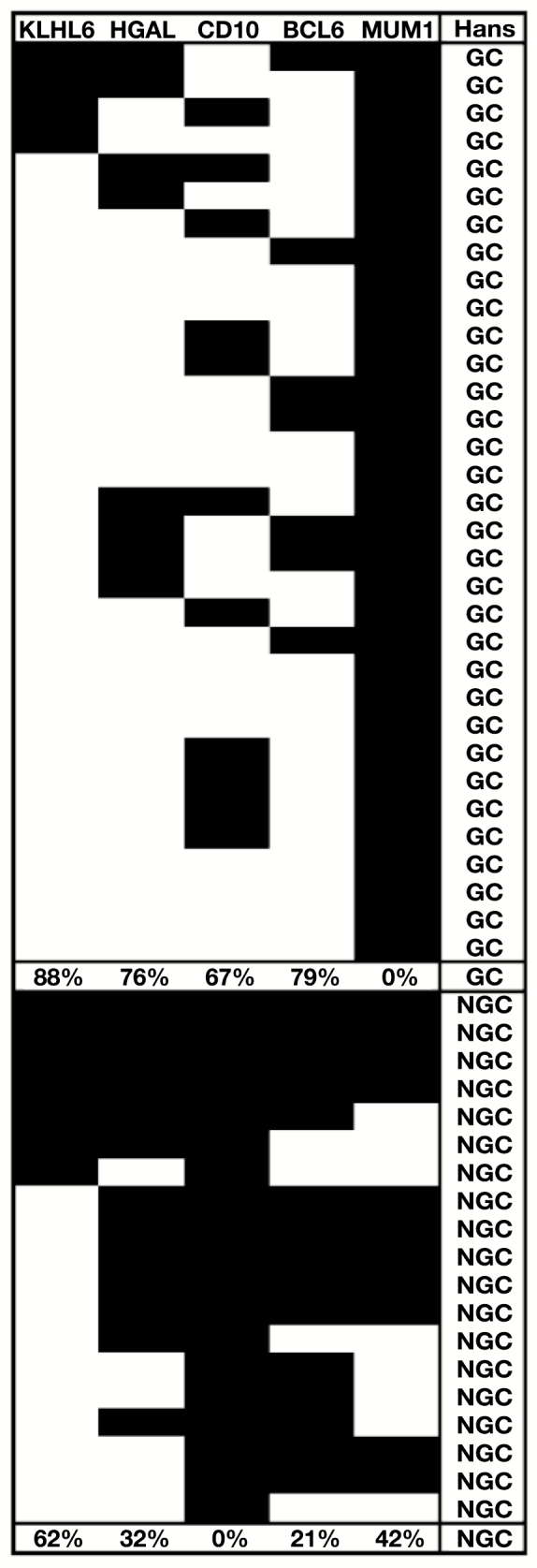
Immunophenotypes of 52 cases of diffuse large B-cell lymphoma. Diagram demonstrating the relative frequencies of germinal center (GC) and Hans et al25 algorithm marker immunophenotypes in 52 cases of diffuse large B-cell lymphoma. The percentage of cases within the Hans-defined GC and non-GC (NGC) groups is given under each column. White indicates marker positivity while black indicates the marker was negative. KLHL6 was considered positive if at least 5% of the cells stained, as described in the Materials and Methods section, whereas cutoffs of 30% were used for the other antibodies, for consistency with the original Hans et al25 model.
To determine the relationship of KLHL6 expression to the GCB and ABC subtypes of DLBCL, a subset of cases was stained for CD10, BCL6, and MUM1, in addition to KLHL6, to determine subtype according to the algorithm by Hans et al.25 Of 325 cases evaluable for all four markers, 173 (53.2%) typed as GCB and 152 (47.8%) typed as ABC according to this algorithm. Of the GCB-type cases, 85.0% were KLHL6 positive. Of the ABC-type cases, 82.2% were KLHL6 positive.
Given the impressive prognostic ability of KLHL6 mRNA expression within DLBCL, we investigated whether protein expression by immunohistochemistry in a cohort of 150 R-CHOP–treated DLBCL cases might also predict prognosis. However, in the context of this cohort and using our conditions for immunohistochemistry, no prognostic significance was observed at the protein level (data not shown).
Discussion
The expression of KLHL6 is restricted to the B-cell lineage and is highly upregulated in B cells during the germinal center reaction.2,9 The protein’s function, however, remains elusive, and homology is not particularly enlightening in this case as BTB-Kelch proteins are involved in a wide variety of cellular activities. The molecule’s possible importance in B-cell function and neoplasia is further supported by the finding that the gene is recurrently mutated in some B-cell lymphomas.11-14
Thus far, there are limited published data on KLHL6 protein expression in hematopoietic malignancies. In Hodgkin lymphoma, Nam-Cha et al26 showed positivity for KLHL6 by immunohistochemistry in 23 of 57 classical Hodgkin lymphomas, 11 of 16 lymphocyte-rich classical Hodgkin lymphomas, and 47 of 60 nodular lymphocyte-predominant Hodgkin lymphomas. A series of plasmablastic lymphomas showed at least weak reactivity in six of 35 cases.27 A study of cutaneous lymphomas showed staining in follicle center cell lymphomas and DLBCL leg type but not in cutaneous marginal zone lymphomas.28
This study complements and expands these early data to a broad variety of human lymphomas with a greater number of cases. It provides additional evidence that KLHL6, at the protein level as well as at the transcription level, is strongly and specifically expressed in germinal center B cells and in hematopoietic neoplasms derived from these cells. This specificity makes KLHL6 a useful diagnostic marker.
Our data show that, like other germinal center markers, KLHL6 is expressed in a subset of DLBCLs. The prognostically significant division of this clinically heterogeneous group of neoplasms into GBC and ABC types was originally based on gene expression profiling,1 but much effort has been expended attempting to devise an immunohistochemical surrogate for this expensive and cumbersome test. The most studied immunohistochemical system for subclassifying cases is by Hans et al,25 who showed that an algorithm using CD10, BCL6, and MUM1 expression could classify cases into groups with significantly different overall survival, independent of the International Prognostic Index, which remains the principal tool for prognostication in DLBCL. This algorithm or subtle variations thereupon have shown prognostic significance in other studies.29,30 More recently, Choi et al31 devised an expanded algorithm (incorporating GCET1 and FOXP1 into the list of markers) and achieved 93% concordance with gene expression profiling performed on the same cases, which was an improvement over the Hans et al25 algorithm alone in their series. Overall, however, the use of immunohistochemistry for stratifying cases of DLBCL into germinal center and non–germinal center types has been difficult to integrate into routine clinical practice, largely because the scoring of some markers, as well as the technical efficacy of staining, may vary widely from laboratory to laboratory.32 In addition, different large studies have shown conflicting results regarding the technique’s value, especially now that essentially all patients with B-cell lymphomas receive rituximab.21,33 Although the current study does not provide an answer to whether addition of another germinal center marker would lead to refinement of immunohistochemistry-based assignment to GCB or ABC types, the observation that most cases typing as both GCB and ABC according to the Hans et al25 algorithm were KLHL6 positive suggests that it may not be.
Nodular lymphocyte-predominant Hodgkin lymphoma is a germinal center B-cell–derived lymphoma with evidence of somatic hypermutation in tumor cells34,35 as well as expression of B-cell and also germinal center B-cell antigens detectable by immunohistochemistry. The situation is somewhat more complex in classical Hodgkin lymphoma, which also shows frequent somatic hypermutation,36 suggesting a post–germinal center B cell of origin. However, most cases do not show high expression of B-cell antigens. The finding that at least a subset of cases expresses KLHL6 provides further evidence to support the germinal center origin of these neoplasms.
Additional highly specific germinal center markers such as KLHL6 are also likely to be useful diagnostic adjuncts in the separation of germinal center and non–germinal center–derived B-cell lymphomas. For example, the distinction of nodal and extranodal marginal zone lymphomas with follicular colonization from follicular lymphoma would be one situation in which additional evidence for germinal center origin may prove helpful.
The observation that KLHL6 mRNA levels predict prognosis in DLBCL is important and deserves further inquiry. In particular, it will be important to determine whether this prognostic power is retained after controlling for known prognostic factors, which was not possible in this study, and a more sophisticated approach for determining the cutoffs between “high” and “low” may also yield a more impressive separation. It is interesting that mRNA levels do not correlate directly with GCB vs ABC subtypes, similar to what was seen with immunohistochemistry, but that prognostic significance was only seen at the mRNA level. This discontinuity is unfortunate but underscores the significant differences between measuring expression at the message and protein levels. It is possible that protein levels reflected in the intensity of staining (reflecting protein abundance) rather than binary absence or presence would be more powerful, as most of our cases were “KLHL6 positive,” whereas for the purposes of the mRNA analysis, half of cases with expression levels below the median was compared with the other half. As such, it remains an open question as to whether prognostic significance can be shown using immunohistochemical intensity as a continuous variable or by altering the cutoff criteria.
It is also notable that mRNA expression was for the most part similar between cases of follicular lymphoma, DLBCL, and Burkitt lymphoma. This is consistent with the finding that the gene is expressed at all stages of B-cell development, but it is surprising that levels were not notably higher in follicular and Burkitt lymphoma, which are both neoplasms of germinal center B cells (the subtype of the DLBCL cases is not known). However, the number of cases was very small, and future studies incorporating more cases and different histologies will better delineate differences in mRNA levels between lymphoma types.
As mentioned above, recurring mutations in KLHL6 have been described in human lymphomas11-14 as well as in other neoplasms, but the distribution of such mutations across lymphoma types has not been reported as yet. We are unaware of the mutation status of KLHL6 in the cases in our sample, but correlation between mutation status and KLHL6 immunohistochemistry would also be a potentially useful avenue for further investigation.
In conclusion, we have investigated the novel germinal center protein KLHL6 in normal and neoplastic tissues at the mRNA and protein levels, and in particular, we have described the spectrum of immunohistochemical staining in lymphomas using a new monoclonal antibody. The specificity of this marker for B cells of germinal center origin makes it a promising tool for the diagnosis of lymphoid neoplasms. Our understanding of the normal germinal center B cell and its neoplastic counterparts continues to evolve with the discovery and characterization of new molecules expressed as part of the germinal center program. New pathologic and molecular insights into this program will likely permit a more biologically and clinically meaningful classification of B-cell lymphomas to emerge in the future.
Supplementary Material
References
- 1. Alizadeh AA, Eisen MB, Davis RE et al. . Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503-511. [DOI] [PubMed] [Google Scholar]
- 2. Gupta-Rossi N, Storck S, Griebel PJ et al. . Specific over-expression of deltex and a new kelch-like protein in human germinal center B cells. Mol Immunol. 2003;39:791-799. [DOI] [PubMed] [Google Scholar]
- 3. Kigoshi Y, Tsuruta F, Chiba T. Ubiquitin ligase activity of cul3-KLHL7 protein is attenuated by autosomal dominant retinitis pigmentosa causative mutation. J Biol Chem. 2011;286:33613-33621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sumara I, Quadroni M, Frei C et al. . A cul3-based E3 ligase removes aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell. 2007;12:887-900. [DOI] [PubMed] [Google Scholar]
- 5. Wakabayashi N, Itoh K, Wakabayashi J et al. . Keap1-null mutation leads to postnatal lethality due to constitutive nrf2 activation. Nat Genet. 2003;35:238-245. [DOI] [PubMed] [Google Scholar]
- 6. Yan W, Ma L, Burns KH et al. . Haploinsufficiency of kelch-like protein homolog 10 causes infertility in male mice. Proc Natl Acad Sci U S A. 2004;101:7793-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bomont P, Cavalier L, Blondeau F et al. . The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26:370-374. [DOI] [PubMed] [Google Scholar]
- 8. Lin Z, Li S, Feng C et al. . Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat Genet. 2016;48:1508-1516. [DOI] [PubMed] [Google Scholar]
- 9. Kroll J, Shi X, Caprioli A et al. . The BTB-kelch protein KLHL6 is involved in B-lymphocyte antigen receptor signaling and germinal center formation. Mol Cell Biol. 2005;25:8531-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Satpathy S, Wagner SA, Beli P et al. . Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol Syst Biol. 2015;11:810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weigert O, Kopp N, Lane AA et al. . Molecular ontogeny of donor-derived follicular lymphomas occurring after hematopoietic cell transplantation. Cancer Discov. 2012;2:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Puente XS, Pinyol M, Quesada V et al. . Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim JA, Hwang B, Park SN et al. . Genomic profile of chronic lymphocytic leukemia in Korea identified by targeted sequencing. PLoS One. 2016;11:e0167641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ganapathi KA, Jobanputra V, Iwamoto F et al. . The genetic landscape of dural marginal zone lymphomas. Oncotarget. 2016;7:43052-43061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krupp M, Marquardt JU, Sahin U et al. . RNA-seq atlas—a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics. 2012;28:1184-1185. [DOI] [PubMed] [Google Scholar]
- 16. Mabbott NA, Baillie JK, Brown H et al. . An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics. 2013;14:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brune V, Tiacci E, Pfeil I et al. . Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med. 2008;205:2251-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lenz G, Wright G, Dave SS et al. ; Lymphoma/Leukemia Molecular Profiling Project Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reich M, Liefeld T, Gould J et al. . Genepattern 2.0. Nat Genet. 2006;38:500-501. [DOI] [PubMed] [Google Scholar]
- 20. Wright G, Tan B, Rosenwald A et al. . A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991-9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Visco C, Li Y, Xu-Monette ZY et al. . Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the international DLBCL rituximab-CHOP consortium program study. Leukemia. 2012;26:2103-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Williams PM, Li R, Johnson NA et al. . A novel method of amplification of FFPET-derived RNA enables accurate disease classification with microarrays. J Mol Diagn. 2010;12:680-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Natkunam Y, Warnke RA, Montgomery K et al. . Analysis of MUM1/IRF4 protein expression using tissue microarrays and immunohistochemistry. Mod Pathol. 2001;14:686-694. [DOI] [PubMed] [Google Scholar]
- 24. Swerdlow SH, Campo E, Pileri SA et al. . The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hans CP, Weisenburger DD, Greiner TC et al. . Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275-282. [DOI] [PubMed] [Google Scholar]
- 26. Nam-Cha SH, Montes-Moreno S, Salcedo MT et al. . Lymphocyte-rich classical Hodgkin’s lymphoma: distinctive tumor and microenvironment markers. Mod Pathol. 2009;22:1006-1015. [DOI] [PubMed] [Google Scholar]
- 27. Montes-Moreno S, Gonzalez-Medina AR, Rodriguez-Pinilla SM et al. . Aggressive large B-cell lymphoma with plasma cell differentiation: immunohistochemical characterization of plasmablastic lymphoma and diffuse large B-cell lymphoma with partial plasmablastic phenotype. Haematologica. 2010;95:1342-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fanoni D, Tavecchio S, Recalcati S et al. . New monoclonal antibodies against B-cell antigens: possible new strategies for diagnosis of primary cutaneous B-cell lymphomas. Immunol Lett. 2011;134:157-160. [DOI] [PubMed] [Google Scholar]
- 29. Berglund M, Thunberg U, Amini RM et al. . Evaluation of immunophenotype in diffuse large B-cell lymphoma and its impact on prognosis. Mod Pathol. 2005;18:1113-1120. [DOI] [PubMed] [Google Scholar]
- 30. van Imhoff GW, Boerma EJ, van der Holt B et al. . Prognostic impact of germinal center-associated proteins and chromosomal breakpoints in poor-risk diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:4135-4142. [DOI] [PubMed] [Google Scholar]
- 31. Choi WW, Weisenburger DD, Greiner TC et al. . A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Jong D, Rosenwald A, Chhanabhai M et al. ; Lunenburg Lymphoma Biomarker Consortium Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications—a study from the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol. 2007;25:805-812. [DOI] [PubMed] [Google Scholar]
- 33. Salles G, de Jong D, Xie W et al. . Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: a study from the Lunenburg Lymphoma Biomarker Consortium. Blood. 2011;117:7070-7078. [DOI] [PubMed] [Google Scholar]
- 34. Braeuninger A, Kuppers R, Strickler JG et al. . Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proc Natl Acad Sci U S A. 1997;94:9337-9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marafioti T, Hummel M, Anagnostopoulos I et al. . Origin of nodular lymphocyte-predominant Hodgkin’s disease from a clonal expansion of highly mutated germinal-center B cells. N Engl J Med. 1997;337:453-458. [DOI] [PubMed] [Google Scholar]
- 36. Kanzler H, Küppers R, Hansmann ML et al. . Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184:1495-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.