Abstract
microRNAs (miRNAs) functioning in gene silencing have been associated with cancer progression. However, common abnormal miRNA expression patterns and their potential roles in cancer have not yet been evaluated. To account for individual differences between patients, we retrieved miRNA sequencing data for 575 patients with both tumor and adjacent nontumorous tissues from 14 cancer types from The Cancer Genome Atlas (TCGA). We then performed differential expression analysis using DESeq2 and edgeR. Results showed that cancer types can be grouped based on the distribution of miRNAs with different expression patterns between tumor and non-tumor samples. We found 81 significantly differentially expressed miRNAs (SDEmiRNAs) in a single cancer. We also found 21 key SDEmiRNAs (nine over-expressed and 12 under-expressed) associated with at least eight cancers each and enriched in more than 60% of patients per cancer, including four newly identified SDEmiRNAs (hsa-mir-4746, hsa-mir-3648, hsa-mir-3687, and hsa-mir-1269a). The downstream effects of these 21 SDEmiRNAs on cellular function were evaluated through enrichment and pathway analysis of 7,186 protein-coding gene targets mined from literature reports of differential expression of miRNAs in cancer. This analysis enables identification of SDEmiRNA functional similarity in cell proliferation control across a wide range of cancers, and assembly of common regulatory networks over cancer-related pathways. These findings were validated by construction of a regulatory network in the PI3K pathway. This study provides evidence for the value of further analysis of SDEmiRNAs as potential biomarkers and therapeutic targets for cancer diagnosis and treatment.
Keywords: miRNA, differential expression analysis, pan-cancer, miRNA target, functional analysis
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs with two forms, the premature miRNA (length 50–125 bp) and the processed, mature form (length 18–24 bp) [1]. miRNAs function in gene silencing by binding to mRNAs, causing either mRNA destabilization or inhibition of translation [2]. From the beginning of the 21st century, pioneer researchers have increasingly catalogued the close relationship between miRNAs and cancer development. For example, Calin et al. [3] discovered the deletion or down-regulation of miR15/miR16 in chronic lymphocytic leukemia, Takamizawa et al. [4] showed under-expressed hsa-let-7 is related to a lower survival rate in lung cancer, Chan et al. [5] knocked down hsa-mir-21 in glioblastoma cells which activated apoptosis, and Li et al. [6] found hsa-mir-10b enabled cell metastasis in breast cancer. These studies not only laid the foundation for a number of studies on the roles of these miRNAs in different cancer types, but also paved the way to the increasing identification of cancerassociated miRNAs [7].
Furthermore, it is believed that there are significantly fewer known human miRNAs (1,881 premature and 2,588 mature in miRBase as of 2016 [1]) than total mRNAs because each miRNA has multiple target mRNAs. Due to this one-to-many relationship, studying miRNA expression profiles could contribute more efficiently to construction of the molecular regulatory network as a whole, especially in characterizing cancer-associated regulation of molecular pathways or biological processes. miRNAs can be classified as either tumor suppressors or oncogenes (oncomirs) according to their expression profiles in cancer [8]. In breast cancer, at least 20 oncogenic miRNAs or cluster families have been identified to promote cell proliferation, invasion, migration, or angiogenesis [9]. Cluster families are groups of several miRNAs transcribed together and co-expressed. Similarly, more than 30 tumor suppressive miRNAs or cluster families have been observed to play a role in cell apoptosis and negative control of cell proliferation or migration [9]. In cancer metastasis, these miRNAs can change the expression of genes essential for cellular homeostasis and for robustness of cell fate decisions, often through various signaling pathways [10]. Therefore, to gain a comprehensive view of the cancer landscape, it is imperative to characterize miRNAs on a large scale and integrate evidence that has been observed across multiple cancer types to build more inclusive regulatory networks.
In large-scale studies, roles of miRNAs have been found to be important in cancer diagnosis, prognosis, and therapy for multiple cancer types [10–12]. Several computational studies have also predicted the networks of miRNAs in human diseases, including cancers, based on either machine learning methods or similarity strategies [13, 14]. Each cancer type has a unique miRNA expression signature that can be classified into different prognostic groups [15]. With an increasing number of studies about cancer incidence, there has been a corresponding increase in observations of tumors with different underlying molecular mechanisms even within a single cancer type [16, 17]. Cancer subtypes can also be identified by miRNA expression profiling, as in the case of non-small cell lung cancer where miRNAs have classified three subtypes by different driving factors (ALK, EGFR, and KRAS) [18]. The diagnostic and prognostic roles of miRNAs further indicate their potential as therapeutic targets and as candidates for clinical trials [19]. Recently, miRNAs have been gradually introduced into personalized medicine approaches for cancer or other disease therapies [20, 21]. Based on the apparent importance of miRNAs in cancer, a number of studies have been conducted on miRNA expression profiles in which statistical analysis have been used to determine changes between disease and control samples to characterize cancer-specific miRNAs in one or two cancers. However, pan-cancer analysis of miRNA expression has not been explored to identify similarity and differences across multiple cancers.
With the introduction of newer, more sensitive procedures in library sample preparation, accompanied by decreasing costs of next-generation sequencing (NGS) methods [22], researchers can now acquire a comprehensive miRNA abundance profiling through deep sequencing [23], despite known complications like low abundant miRNAs and miRNA processing complexity. Several normalization methods for miRNA sequencing (miRNA-seq) data have been widely used in previous studies [24, 25], including edgeR (Trimmed Mean of M values, TMM) [26] and DESeq [27]. In this study, we performed differential expression analysis by both DESeq2 [28] and edgeR [26] for paired miRNA-seq data (tumor and surrounding nontumorous tissues) from The Cancer Genome Atlas (TCGA: http://cancergenome.nih.gov;https://portal.gdc.cancer.gov/), following the methods described by Yang S et al. [29] and used in the study of Metpally RP et al. [22].
Although previous studies have demonstrated critical roles of miRNAs in the prognosis of cancer types and subtypes, our research singled out key miRNAs observed to have significantly differential expression (SDEmiRNAs) for 14 TCGA cancer types independently. The distribution of these SDEmiRNAs with different expression trends enables us to group the 14 cancers and identify different potential developmental mechanisms of each cancer.Comparing the expression profiles of these SDEmiRNAs in individual cancer types, we were able to compile a list of miRNAs which are significantly differentially expressed in individual cancers. These unique SDEmiRNAs specific to one cancer could be considered biomarkers of each respective cancer, and may influence patient survival rate, suggesting roles as drivers. We also identified a subset of key SDEmiRNAs that are significantly over- or under-expressed with high patient frequencies in at least eight cancer types (over-expression implies functional up-regulation). From the functional and enrichment analyses of their cancer-related target genes and pathways, we find that these SDEmiRNAs might be common functional and executive factors promoting cancer development via similar molecular mechanisms across multiple cancers. As an example, we constructed common regulatory networks of several SDEmiRNAs in signaling pathways across different cancer types, despite great differences among their target genes in multiple tissues. Notably, our study proposes that some identified SDEmiRNAs that are barely studied in cancer development due to their relatively low expression levels have been found to be significantly differentially expressed in a wide range of cancer types, showing their potential to play vital roles in cancer development.
Thus, this study evaluates the expression profiles of human miRNAs in 14 cancer types, investigates the roles of individual SDEmiRNAs to cancer types where they have not previously been studied, proposes potential driver miRNAs in both individual and multiple cancers, classifies the types of functions of key SDEmiRNAs in different tissues according to a narrow subset of cellular molecular mechanisms, and, therefore, provides a means for better discovery of cancer development mechanisms that could improve downstream clinical studies by prioritizing miRNA and targets for translational study. Results are integrated in the open-source BioXpress v3.0 (https://hive.biochemistry.gwu.edu/bioxpress) to support further analysis.
Results and Discussion
Data collection and overall evaluation before differential expression analysis
The TCGA project provides a comprehensive database with a unified data analysis pipeline for collecting and analyzing samples. We were able to collect tumor-only expression data for 1,904 miRNAs in 32 TCGA/ICGC cancer types generated by the HiSeq platform (7,994 TCGA patients and 168 ICGC patients) and 1,668 miRNAs in 12 TCGA cancer types generated by Genome Analyzer (1,439 TCGA patients), from a total of 9,601 patients. From these datasets, 22 TCGA cancer types contained paired data (tumor data with matched adjacent non-tumorous data) from the HiSeq platform: we set a threshold for patient counts (greater than 10, covering more than 95% of total patients with paired data), leaving 14 cancer types to be used in further analysis (BLCA, BRCA, PRAD, STAD, KIRP, KIRC, KICH, LUAD, LUSC, LIHC, THCA, ESCA, UCEC, and HNSC – please see Table 1 for full cancer names). These cancer types included a total of 575 TCGA-only patients (1,152 paired samples) with an average of 1,393 expressed miRNAs for each cancer.
Table 1.
Summary of patients and miRNA expression counts, and significantly differentially expressed miRNAs over- and under-expressed counts reported by DESeq2 and edgeR for paired data of 14 TCGA cancer types
| TCGA-Cancer Type | TCGA Cancer Type (full name) | Mapped Cancer Type (DOID) | Patient Counts | Expressed miRNA Counts | miRNA sig over-expressed | miRNA sig under-expressed | Total sig miRNA | 80% patients* |
|---|---|---|---|---|---|---|---|---|
| BLCA | Bladder Urothelial Carcinoma | DOID:11054 / Urinary bladder cancer [UBC] | 19 | 1361 | 18 | 115 | 133 | 72 |
| UCEC | Uterine Corpus Endometrial Carcinoma | DOID:363 / Uterine cancer [Uteca] | 18 | 1370 | 39 | 193 | 232 | 115 |
| LIHC | Liver hepatocellular carcinoma | DOID:3571 / Liver cancer [Livca] | 48 | 1414 | 98 | 70 | 168 | 52 |
| HNSC | Head and Neck squamous cell carcinoma | DOID:11934 / Head and neck cancer [H&NC] | 43 | 1463 | 100 | 152 | 252 | 65 |
| STAD | Stomach adenocarcinoma | DOID:10534 / Stomach cancer [Stoca] | 41 | 1426 | 28 | 126 | 154 | 51 |
| ESCA | Esophageal carcinoma | DOID:5041 / Esophageal cancer [EC] | 12 | 1215 | 4 | 38 | 42 | 24 |
| THCA | Thyroid carcinoma | DOID:1781 / Thyroid cancer [Thyca] | 59 | 1506 | 50 | 116 | 166 | 59 |
| PRAD | Prostate adenocarcinoma | DOID:10283 / Prostate cancer [PCa] | 52 | 1350 | 16 | 64 | 80 | 21 |
| BRCA | Breast invasive carcinoma | DOID:1612 / Breast cancer [BRCA] | 75 | 1445 | 123 | 163 | 286 | 58 |
| LUSC | Lung squamous cell carcinoma | DOID:1324 / Lung cancer [Lunca] & DOID:3907 / Lung squamous cell carcinoma | 44 | 1468 | 89 | 205 | 294 | 74 |
| LUAD | Lung adenocarcinoma | DOID:1324 / Lung cancer [Lunca] & DOID:3910 / Lung adenocarcinoma | 39 | 1403 | 63 | 164 | 227 | 91 |
| KICH | Kidney Chromophobe | DOID:263 / Kidney cancer [Kidca] & DOID:4471 / Chromophobe adenocarcinoma | 24 | 1312 | 148 | 67 | 215 | 119 |
| KIRC | Kidney renal clear cell carcinoma | DOID:263 / Kidney cancer [Kidca] & Kidney renal clear cell carcinoma | 67 | 1363 | 133 | 134 | 267 | 74 |
| KIRP | Kidney renal papillary cellcarcinoma | DOID:263 / Kidney cancer [Kidca] & DOID:4465 / Papillary renal cell carcinoma | 34 | 1400 | 135 | 99 | 234 | 85 |
Note:
80% patients: counts of miRNAs that are significantly over-expressed (significantly under-expressed) in at least 80% of patients in each cancer type.
To obtain an overview of the expression profiles of miRNAs in paired samples from the 14 cancer types, Reads Per Million miRNA (RPM) values of each miRNA (provided by TCGA) in each TCGA patient were first used to calculate regular fold change (FC) values between cancer and corresponding non-tumorous samples. After comparing these FC values, we found that the total number of over-expressed miRNAs is two-fold higher than that of under-expressed miRNAs. However, when considering only those miRNAs with FC > 2 for over-expression and FC < 0.5 for under-expression, and with patient frequencies > 35% in at least eight cancers, the distribution of miRNAs with over- and under-expression are similar to each other in each cancer (Figure 1). This suggests that many moderately over-expressed miRNAs could be effectors driven by other factors and may vary by patient.
Figure 1. Overview of miRNA counts vs. patient frequency for over- and under-expressed miRNAs with original FC > 2 of FC < 0.5 in at least eight cancer types from paired data.
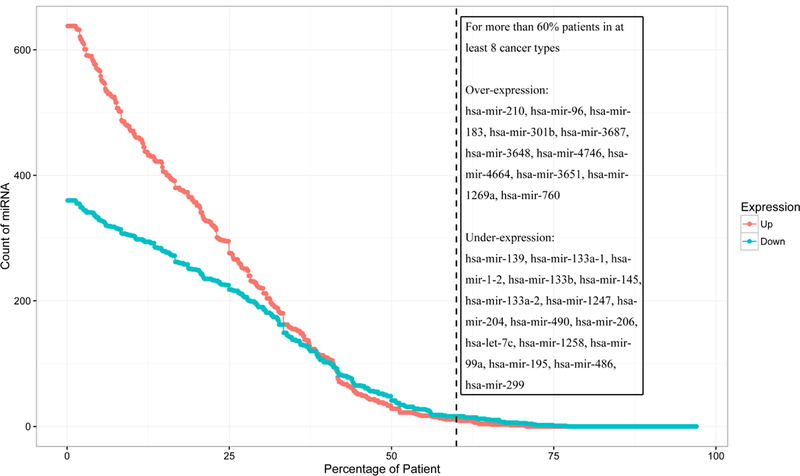
FC is used to distinguish the overall expression trend of miRNAs (by comparing FC values from tumor and non-tumor samples from the same patient). Each dot on the red line represents the count of over-expressed miRNAs per patient frequency, and dots on the blue line represent counts of under-expressed miRNAs. Across more than 35% of patients, the numbers of miRNAs with over- and under-expression are similar to each other. 11 over-expressed miRNAs and 16 under-expressed miRNAs appear in more than 60% of patients for at least eight cancer types: these miRNAs are listed in the text box. These miRNAs in multiple cancer types suggest their potential roles in the development of these cancers.
This expression distribution also highlights 27 miRNAs (11 over-expressed miRNAs and 16 under-expressed miRNAs) that are found in more than 60% of patients (Figure 1) in eight or more cancers. Most of these miRNAs have been previously reported in at least one publication to be associated with cancer, some of which are well-studied tumor suppressors, including hasmir-133b, and hsa-mir-139, or contribute to cell invasion, such as hsa-mir-301b [30, 31]. Notably, hsa-mir-1247 was recently found to be a prognostic marker in pancreatic cancer as a suppressor [30] and hsa-mir-1258 targets heparanase (HPSE) to suppress breast and nonsmall cell lung cancer [32, 33]. hsa-mir-1269a, hsa-mir-3687, and hsa-mir-3648 were also recently identified to be involved in cancer development [34–36], so their molecular mechanisms in cancer have yet to be thoroughly researched. Additionally, two miRNAs reported in 2011 [37], hsa-mir-4746 and hsa-mir-4664, were over-expressed in 11 and eight cancers with average patient frequencies of 74.42% (average FC = 3.95) and 75.49% (average FC = 8.07), respectively, although their average expression levels are lower than many well-studied miRNAs, which may lead to difficulties in quantification and qualification.
Our data are consistent with previous studies on individual miRNAs in cancers, validating the reliability and confidence of our data, and suggesting potential similar roles of the subset of 27 miRNAs found in development of multiple cancers, based on their similar expression profiles. It is also important to note that 21 of these 27 miRNAs were reported to be significantly differentially expressed (SDEmiRNAs) in the same cancer types by the statistical analysis described in the following sections,.
miRNA differential expression analysis
SDEmiRNAs could be used to distinguish cancer types
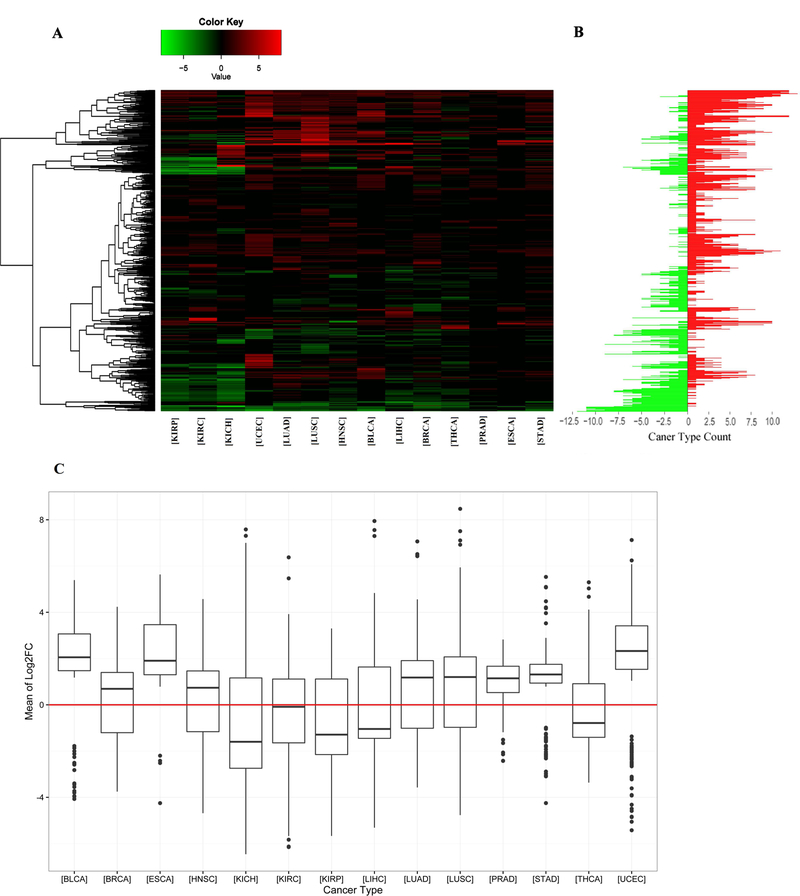
Previous studies have proposed multiple methods to identify tissue-specific miRNAs in distinguishing cancer primary origins, such as feature selection strategies introduced by Tang et al. [38]. However, as more stable and mature tools, DESeq2 [28] and edgeR [26] have been prevalent in expression research [39]. In this study, we utilized both DESeq2 and edgeR to perform miRNA differential expression analysis in 14 cancer types, applying statistical tests to minimize potential false positive errors resulting from the differences among individuals and samples. Data from all patients for a single cancer type were analyzed together resulting in reporting average differential expression levels by cancer type. Both DESeq2 and edgeR provide p values and adjusted p values (or FDR in edgeR). The results of the two tools showed a similar ranking of significantly differentially expressed miRNAs (SDEmiRNAs) when ranked by adjusted p values. We found 10 to 15% of miRNAs with significant differential expression in each cancer type reported by both DESeq2 and edgeR (Table 1). As shown in Figure 2A, of all 1,870 miRNAs in our integrated database, the specific distributions of the quantities and types of the 656 SDEmiRNAs vary between cancer types, suggesting the existence of unique SDEmiRNA patterns for each of these cancer types. These unique patterns of SDEmiRNAs could potentially reduce the amount of miRNAs required for prognosis of cancer types based on the findings of Lu et al. [15]. The results of differential expression analysis of each miRNA is available in BioXpress, shown by both figures and table (Figure S1).
Figure 2. Differential expression analysis of miRNAs and distribution of log2FC values of significantly differentially expressed miRNAs across 14 cancer types.
A. Heatmap showing significantly over- and under-expressed miRNAs in at least one cancer type - vertical axis is miRNAs. The expression change of miRNA is represented by log2FC values generated by DESeq2 and edgeR. Log2FC values less than zero are considered as under-expressed miRNAs (green), while log2FC greater than zero represents over-expressed miRNAs (red). The miRNA patterns of each cancer type vary from each other. B. Counts of cancer types in which each miRNA is over- or under-expressed - vertical axis stands miRNAs (same as Figure 2A). Green bars represent the counts of cancer types in which a miRNA is under-expressed, and red ones are for cancer type counts in which a miRNA is over-expressed. After unsupervised clustering, some miRNAs show the same trend of over-expression or underexpression across most cancers. It suggests that these miRNAs may have similar function across different cancer types. C. The vertical axis represents the average of log2FC values generated from DESeq2 and edgeR for each miRNA in each cancer type. For each cancer, all of its miRNAs that are significantly differential expressed are included. The numbers of over- and under-expressed miRNAs per cancer vary from each other, suggesting the up-regulation of oncogenes is more important in the occurrence of some cancers, while the down-regulation of tumor suppression genes is more important in other cancers.
Moreover, similar SDEmiRNA expression patterns are shown in LUAD and LUSC from lung tissue, as well as in KICH, KIRP, and KIRC from kidney tissue, as expected. However, among 387 SDEmiRNAs in either LUAD or LUSC (Figure S2A), 190 have log2FC values greater than 1 (∆log2FC > 1), 140 (36.2% of 387) of which have the opposite expression trend for both cancers, suggesting they could be key factors underscoring the differences between LUAD and LUSC. We performed a similar pairwise comparison among KICH, KIRP, and KIRC using the set of 330 SDEmiRNAs with ∆log2FC > 1 in at least one of the three cancer types. Findings show that KIRP and KIRC have more similar SDEmiRNA expression patterns than KIRP and KICH, and KICH and KIRC (Figure S2B-D). To the best of our knowledge, this is the first time that analysis suggests the development of KICH could be mechanistically more distant than the development of KIRC and KIRP.
Expression patterns of SDEmiRNAs group cancer types based on different mechanisms during cancer development
We counted the number of SDEmiRNAs with the same expression trend in at least 80% of patients (by log2FC values) for each cancer type (Table 1, and the corresponding patient frequencies in Figure S3). Results showed different cancer types have different proportions of SDEmiRNAs: some cancers have more over-expressed SDEmiRNAs than under-expressed; some have a balance between over- and under-expressed SDEmiRNAs; and other cancer types have more under-expressed SDEmiRNAs (Figure S3). Similar results were obtained when the log2FC values of all SDEmiRNAs were used to display their distributions in all cancer types (Figure 2C).
In four cancer types (KICH, KIRP, LIHC, and THCA), the numbers of under-expressed SDEmiRNAs are much higher than those of over-expressed SDEmiRNAs while, for most of the cancer types (BLCA, ESCA, KIRC, LUAD, LUSC, PRAD, STAD, and UCEC), the median expression values of SDEmiRNAs are greater than zero. Because the main function of miRNA is to suppress target genes, the significant number of under-expressed miRNAs corresponds to the up-regulation of oncogenes, which may play a more important role in the occurrence and progression of the four cancers characterized predominantly by under-expression of miRNAs. Conversely, the down-regulation of tumor suppression genes might be more important in cancer occurrence and progression for those cancers characterized predominantly by over-expression of miRNAs. Cancers like BRCA, HNSC, and KIRC have an almost symmetric distribution of SDEmiRNAs about log2FC = 0 (Figure 2C), suggesting regulation of both oncogenes and tumor suppression genes could be equally important for disease progression. However, it could also be possible that the similar distribution between over- and under-expressed miRNAs indicates that these miRNAs are equally unimportant to disease and may suggest other mechanisms are primarily at work in these cancer types.
Although there are distinguishable differences in development patterns in different types of cancer, our results suggest that the distribution of SDEmiRNAs can group cancers into one of three categories based on the distribution of miRNA expression changes and related regulatory mechanisms employed by each group during cancer initiation and progression. We propose application of these findings will improve efficiency of future research on exploring mechanisms of cancers by organizing studies by group instead of individual cancer types.
SDEmiRNAs unique to one of the 14 cancer types
Although we are mostly interested in SDEmiRNAs implicated in multiple cancers (as discussed in depth below), SDEmiRNAs that only exist in a single cancer type could be biomarkers specific for that cancer type. To investigate this potential, we set up screening conditions to identify those SDEmiRNAs affecting a single cancer type: 1) SDEmiRNA should be reported to be significant by both DESeq2 and edgeR for a given cancer type; 2) SDEmiRNA should not exist in the remaining 13 cancers; 3) the patient frequency for this SDEmiRNA should be greater than 50% in this cancer; 4) the log2FC value for this SDEmiRNA should be greater than 1 or smaller than −1.
As shown in Table S1, we identified a total of 81 SDEmiRNAs with significant expression trends in a single cancer type, 57 of which have patient frequencies higher than 60% (Table 2). Several of these SDEmiRNAs have much lower expression in tissues compared with previously well-studied miRNAs, and have not been functionally studied in cancer development, especially pertaining to their roles in the corresponding cancer types reported in this study. However, this does not negate their potential cancer-related functions. In order to further explore the results, as an example, we performed survival analysis on four SDEmiRNAs unique to BRCA (absolute log2FC values > 1) by using miRpower [40]. Except hsa-mir-329–2 which is not in miRpower, the remaining three SDEmiRNAs suggest significant survival differences (log-rank p value < 0.01) between groups with under-/over-expression of each SDEmiRNA after optimizing the patient threshold (Figure S4): hsa-mir-4784 (Hazard Ratio (HR) = 1.82), hsa-mir-1262 (HR = 0.58), and hsa-mir-320c-1 (HR = 0.68). The differences of survival rates between patients with under- and over-expression were consistent with our results. Such miRNAs should be prioritized for further analysis.
Table 2.
Unique miRNAs with significant differential expression in 60% or more patients of individual cancer
| TCGA type | cancer | Unique miRNAs | Expression trend |
|---|---|---|---|
| BLCA | hsa-mir-3913-1, hsa-mir-6728, hsa-mir-520b, hsa-mir-4523 | Up | |
| BRCA* | hsa-mir-4784 | Up | |
| hsa-mir-605, hsa-mir-320c-1, hsa-mir-1262, hsa-mir-329-2 | Down | ||
| ESCA | hsa-mir-6715b | Up | |
| HNSC | hsa-mir-1305 | Up | |
| hsa-mir-378i, hsa-mir-499a | Down | ||
| KICH | hsa-mir-3152, hsa-mir-7641-2, hsa-mir-582, hsa-mir-548j | Up | |
| hsa-mir-4649, hsa-mir-6761, hsa-mir-3154 | Down | ||
| KIRC | hsa-mir-122, hsa-mir-4773-2, hsa-mir-599, hsa-mir-7110 | Up | |
| KIRP | hsa-mir-3189, hsa-mir-6843, hsa-mir-3680-1, hsa-mir-4676 | Up | |
| hsa-mir-380 | Down | ||
| LIHC | hsa-mir-5187 | Up | |
| hsa-mir-6503, hsa-mir-5701-1 | Down | ||
| LUAD | hsa-mir-372, hsa-mir-4728, hsa-mir-5091 | Up | |
| LUSC | hsa-mir-4665, hsa-mir-4766, hsa-mir-1248 | Up | |
| hsa-mir-1294, hsa-mir-3920, hsa-mir-4423, hsa-mir-5680, hsa-mir-3926-2, hsa-mir-3195 | Down | ||
| PRAD | hsa-mir-5694, hsa-mir-5706 | Up | |
| STAD | hsa-mir-551a, hsa-mir-5703, hsa-mir-3194 | Up | |
| THCA | hsa-mir-6730 | Up | |
| hsa-mir-548q, hsa-mir-876, hsa-mir-1179 | Down | ||
| UCEC | hsa-mir-3131, hsa-mir-1914, hsa-mir-1276, hsa-mir-4763, hsa-mir-3616, hsa-mir- 3187, hsa-mir-5581, hsa-mir-3193 | Up | |
| hsa-mir-548aw | Down |
Pan-cancer analysis of key SDEmiRNAs
As shown in Figure 2B, after unsupervised clustering of all SDEmiRNAs, several SDEmiRNAs follow the same trend of either over-expression or under-expression across most of the cancer types, suggesting that these SDEmiRNAs may function similarly across different cancer types. To further study the key SDEmiRNAs affecting multiple cancers, we identified a subset of 90 miRNAs that have significant differential expression in at least eight cancer types (58% of total cancer types).
Selected SDEmiRNAs were verified against experimentally validated databases, text mining , and machine-learning-based prediction databases
Before proceeding with the analysis, we first confirmed these 90 key SDEmiRNAs generated from DESeq2 and edgeR using four experimentally validated databases. Among 2,280 significantly differentially expressed miRNA-cancer correlations, 208 (9.12%) correlations were recorded in the union of the four databases (HMDD, Mir2disease, miRCancer, and miRiaD) to be exclusively over- or under-expressed in certain types of cancer. Furthermore, we randomly selected 24 of the 90 SDEmiRNAs (with a total of 226 miRNA-cancer interactions) as the input for a text mining tool (DEXTER) [41]. After mapping and manual curation, we extracted 157 (69.47%) interactions (Table S2). None of the 24 SDEmiRNAs were identified by DEXTER to have opposite expression trends as compared to the trends we reported in our study.
Notably, from the 90 selected SDEmiRNAs, we defined a subset based on conservation of theirdifferential expression trend across cancer types: 47 SDEmiRNAs are all over-expressed and 18 are all under-expressed in eight or more cancer types (a total of 65 SDEmiRNAs with the same significant differential expression trend in eight or more cancers) (Table S3). We compared these results with two miRNA-disease prediction databases: Infer microRNA-disease association (http://server.malab.cn:50001) [42] and MicroRNA-Disease Association Prediction (MDAP) (http://server.malab.cn/MDAP) [43]. In 466 unique associations in Table S3 (considering only mature miRNA sequences), except 21 miRNAs (with 185 associations) which were identified after publication of these two databases, 163 of the remaining 281 associations (58%) were confirmed by the two prediction databases.
Key SDEmiRNAs with at least 60% patient frequencies were considered reliable and may have similar functions across multiple cancers
Of 65 SDEmiRNAs that have the same significant expression change trend in eight or more cancers (Table S3), 54 SDEmiRNAs belong to 40 miRNA families, according to MirGeneDB v2.0 [44]. Among them, all members of 22 families are included in the list, although most of the families have only one or two members. However, the similar expression trends of these SDEmiRNAs suggest their high consistency of activities and potential functions across 58% of investigated cancers, and therefore, these small families can be classified into larger ones (Table S3). Individually, six newly reported miRNAs were included in the 65 SDEmiRNAs: hsamir-3170, hsa-mir-3677, hsa-mir-4326, hsa-mir-4652, hsa-mir-7706, and hsa-mir-105–2 (Table S3). Among them, hsa-mir-4652 is the top significantly over-expressed miRNA with largest log2FC value (average log2FC = 5.34, patient frequency larger than 60% in seven cancer types). We found no cancer-related studies that have explored the mechanisms of these six miRNAs in cancers, suggesting their high potential for involvement in cancer development, especially hsa-mir-4652.
Comparing the 65 SDEmiRNAs identified by differential expression analysis to the 27 miRNAs with regular FC > 2 or FC < 0.5 across 60% of patients (in ‘Data collection and evaluation’ section, Figure 1) in at least eight cancer types, we found an overlap of nine over-expressed and 12 under-expressed SDEmiRNAs between the two datasets (a total of 21 miRNAs) (Table 3). This subset not only includes well-studied miRNAs such as hsa-mir-145 and hsa-mir-210 reported in previous publications, but also newly identified miRNAs such as hsa-mir-4746 (in ‘Data collection and evaluation’ section), hsa-mir-3648, hsa-mir-3687, and hsa-mir-1269a. Two of the over-expressed and five of the under-expressed key miRNAs are in the top five significantly over- and under-expressed miRNAs of the 65 ranked by average log2FC values (Table 3).
Table 3.
21 miRNAs with significant differential expression and high patient frequencies (at least 60%) in tumor samples from each of eight or more cancers compared to non-tumorous samples
| miRNAa | Cancer type | Mean log2FC | Mean freq (strFC)b | Expression change |
|---|---|---|---|---|
| hsa-mir-1269a | LUSC, LUAD, LIHC, UCEC, STAD, BRCA, HNSC, PRAD | 5.18 | 64.58% | Up |
| hsa-mir-210 | LUSC, LUAD, BLCA, KIRC, UCEC, BRCA, HNSC, KIRP | 3.68 | 48.13% | Up |
| hsa-mir-301b | UCEC, LUSC, BLCA, LUAD, BRCA, ESCA, HNSC, KIRC, STAD, KIRP | 2.71 | 56.53% | Up |
| hsa-mir-183 | UCEC, BLCA, LIHC, KICH, BRCA, LUSC, LUAD, STAD, KIRP, PRAD, HNSC, THCA | 2.68 | 48.13% | Up |
| hsa-mir-3648 | LUSC, UCEC, ESCA, LUAD, BLCA, HNSC, PRAD, STAD, BRCA | 2.57 | 52.53% | Up |
| hsa-mir-3687 | BLCA, LUSC, LUAD, STAD, UCEC, HNSC, BRCA, PRAD | 2.56 | 52.91% | Up |
| hsa-mir-96 | UCEC, BLCA, KICH, BRCA, LIHC, LUSC, LUAD, STAD, PRAD, KIRP, THCA, HNSC | 2.56 | 49.69% | Up |
| hsa-mir-760 | LUSC, LIHC, LUAD, KIRC, KIRP, BRCA, UCEC, KICH, HNSC | 2.02 | 37.31% | Up |
| hsa-mir-4746 | UCEC, LIHC, ESCA, LUSC, KIRP, BRCA, HNSC, BLCA,KIRC, STAD, LUAD | 1.96 | 35.94% | Up |
| hsa-mir-206 | BRCA, THCA, LUAD, KIRP, KIRC, LUSC, UCEC, KICH | −4.18 | 75.38% | Down |
| hsa-mir-204 | KIRP, PRAD, LUAD, KIRC, STAD, THCA, HNSC, BLCA, BRCA, ESCA, KICH | −2.78 | 56.47% | Down |
| hsa-mir-1-2 | KIRC, BRCA, KIRP, KICH, HNSC, ESCA, LUAD, LUSC, STAD, BLCA, UCEC | −2.74 | 59.78% | Down |
| hsa-mir-133a-2 | PRAD, KIRC, LUAD, LUSC, KIRP, KICH, STAD, BLCA, UCEC | −2.72 | 59.94% | Down |
| hsa-mir-133b | THCA, PRAD, HNSC, KIRC, LUAD, KIRP, LUSC, KICH, STAD, BLCA, UCEC | −2.65 | 57.66% | Down |
| hsa-mir-133a-1 | THCA, KIRC, BRCA, HNSC, KIRP, ESCA, LUAD, KICH, LUSC, STAD, BLCA, UCEC | −2.6 | 59.94% | Down |
| hsa-mir-1247 | KIRC, LUSC, HNSC, LIHC, LUAD, THCA, KIRP, BRCA, KICH, BLCA, UCEC | −2.38 | 56.31% | Down |
| hsa-mir-139 | KIRC, THCA, LUAD, LIHC, HNSC, STAD, KIRP, LUSC, BRCA, BLCA, UCEC | −2.21 | 55.01% | Down |
| hsa-mir-145 | HNSC, THCA, LUAD, LIHC, LUSC, KICH, BRCA, STAD, KIRP, BLCA, UCEC | −1.73 | 39.98% | Down |
| hsa-mir-195 | LIHC, THCA, STAD, KICH, BRCA, HNSC, LUSC, LUAD, KIRP, BLCA, UCEC | −1.7 | 39.91% | Down |
| hsa-mir-99a | THCA, KICH, LIHC, LUAD, LUSC, BRCA, HNSC, UCEC | −1.63 | 33.44% | Down |
| hsa-let-7c | THCA, LIHC, KICH, LUSC, LUAD, HNSC, BRCA, UCEC | −1.55 | 34.02% | Down |
Molecular mechanisms for the newly identified hsa-mir-1269a have rarely been published for the cancer types we report here, but Min et al. have recently found a single-nucleotide variation in hsa-mir-1269a that could contribute to the occurrence of hepatocellular carcinoma [45]. In our results, hsa-mir-1269a has log2FC value consistently larger than 2.20 in each of its eight associated cancer types (average log2FC = 5.18), with the average frequency of affected patients greater than 64.58% for a strict fold change requirement (FC > 5 for each patient). Other members of the 21 key SDEmiRNAs (Table 3) include hsa-mir-183 and hsa-mir-96, belonging to the mir-183-96-182 cluster, which functions to induce cell proliferation and cancer development [46]. Notably, unlike the other two miRNAs from the same cluster, hsa-mir-182 only shows significant differential expression but not high patient frequencies. Considering our relatively stringent criteria on FC values (FC > 2 or FC < 0.5), the changes of hsa-mir-182 in most patients are not as strong as those of hsa-mir-183 and hsa-mir-96.
Overall, our study groups 14 cancer types with potential different molecular mechanisms of development based on the distribution of SDEmiRNAs, as well as proposes that some SDEmiRNAs previously indicated to play a role in distinct cancer types may function similarly in the development of additional cancer types, and classifies certain small miRNA families into larger ones. Moreover, the results correspond well with those of previously published studies and include evaluation of the 21 key SDEmiRNAs with high patient frequencies in a comprehensive pan-cancer analysis. Newly identified miRNAs hsa-mir-4746, hsa-mir-3648, hsamir-3687, and hsa-mir-1269a could also be important factors in multiple cancer types, although their relations to cancer were rarely reported by previous studies due to their low expression levels compared to others.
SDEmiRNA targets enrichment and functional analysis
Each of the 65 SDEmiRNAs discussed above (with the same significant differential expression trend in eight or more cancer types) participates in some form of molecular regulation, either inducing or inhibiting cell proliferation and cancer metastasis in different cancer types. To better understand the general pathways or biological processes by which SDEmiRNAs contribute to the occurrence and development of multiple cancer types, we extracted experimentally validated non-tissue-specific targets for those SDEmiRNAs without tissue or cancer information, as well as tissue-specific targets.
Distribution and functional analysis of non-tissue-specific targets of 65 SDEmiRNAs
60 of 65 selected miRNAs were found to have a total 9,896 targets with a total of 29,087 miRNA-target interactions (the other five miRNAs had no available target information). Among the target genes, 901 genes are considered to be cancer census genes from COSMIC or cancer biomarkers from EDRN, corresponding to 58 miRNAs. Cancer census genes are those genes with substantial evidence for their relations to cancer from published research [47]. hsa-mir-21 alone regulates 471 cancer-related genes, and hsa-mir-93 regulates 234 cancer-related genes. We then identified a subset of 27 cancer-related genes that are targets of more than 10 miRNAs, and among all miRNAs involved in regulating these 27 genes, 36 are found that target four or more. As shown in Figure 3, some of the cancer-related genes, such as CCND2 (G1/Sspecific cyclin-D2) (oncogene), MDM2 (E3 ubiquitin-protein ligase Mdm2) (oncogene), CDKN1A (Cyclin-dependent kinase inhibitor 1) (tumor suppressor), and SMAD4 (Mothers against decapentaplegic homolog 4) (tumor suppressor), could be regulated by multiple miRNAs and the numbers of targets of hsa-mir-106b, hsa-mir-19a, hsa-mir-195, hsa-mir-130b, and hsa-mir21 are greater than that of other miRNAs with identified targets.
Figure 3. Non-tissue-specific cancer-related genes which are targets of more than 10 miRNA types.
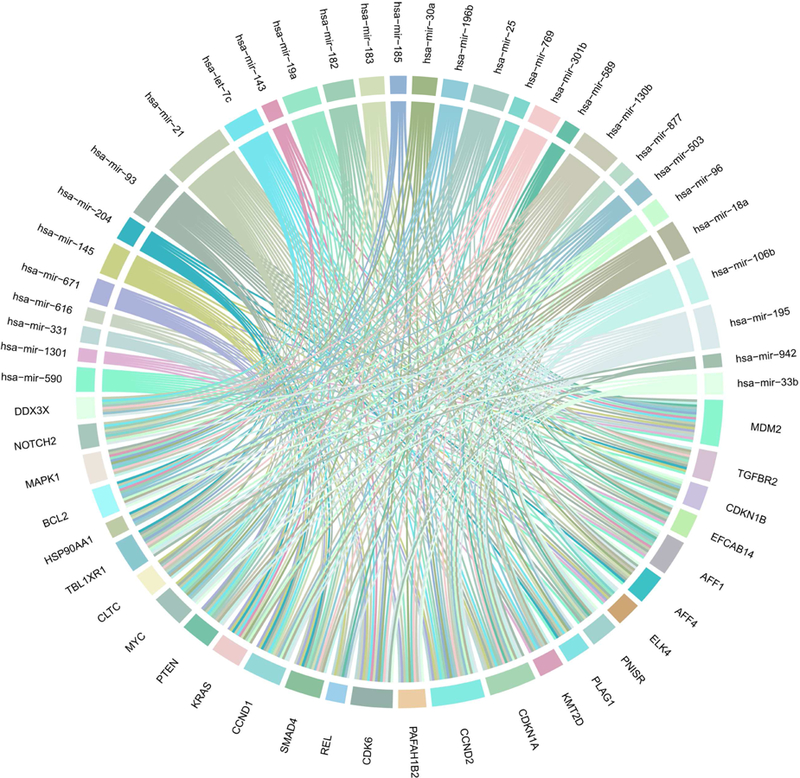
Each unique color represents one miRNA with at least four targets and the ribbon width suggests the interactions between the miRNAs and target genes. 27 cancer-related genes are targets of more than 10 miRNAs, and among all miRNAs involved in regulating these 27 genes, 36 target four or more cancer-related genes. Some of the cancer-related genes could be regulated by multiple miRNAs, and the numbers of targets of some miRNAs are greater than those of others. These miRNAs play vital roles in cancer progression via regulating cancer-related genes.
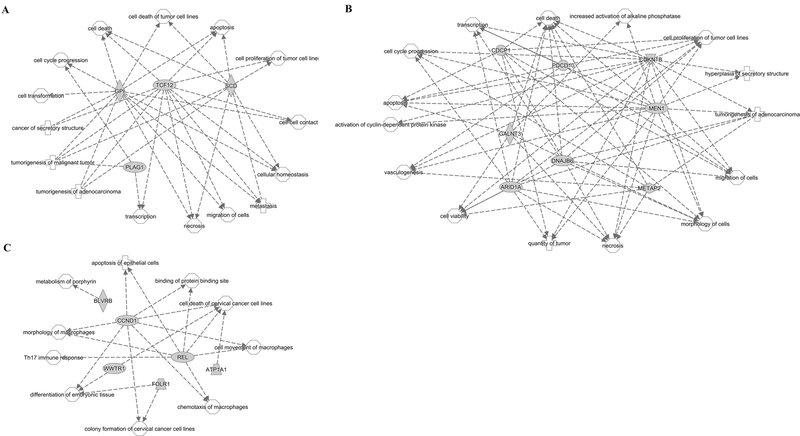
For the 21 key SDEmiRNAs with high patient frequencies (FC > 2 or < 0.5) (Table 3), in addition to hsa-mir-4652, which has the largest log2FC values and greater than 60% patient frequency in seven cancers, 18 were found to have 428 cancer-related genes out of their 3,987 non-tissue-specific targets. Fifteen of the 21 key SDEmiRNAs regulate 25 cancer-related genes. Eleven of these SDEmiRNAs function in enzyme binding (p value = 8.30e-03, by STRING), lending credence to their regulatory roles in cancer development. Well-studied miRNAs such as hsa-mir-195, hsa-let-7c, and hsa-mir-183 have more than 70 cancer-related targets each. In particular, hsa-mir-1269a has 57 (five cancer-related), hsa-mir-4652 has 133 (12 cancerrelated), hsa-mir-4746 has 54 (six cancer-related), hsa-mir-3648 has 14 (one cancer-related), and hsa-mir-3687 has 14 (no cancer-related) non-tissue-specific targets. Our analysis by Ingenuity Pathway Analysis (IPA) suggests these targets participate in various important networks and cellular processes (Table 4). For cancer-related targets of hsa-mir-1269a and hsamir-4652, most are involved in cell survival, such as cell cycle progression, cell migration, and apoptosis (Figure 4A-B), while the functions of targets of hsa-mir-4746 are less uniform and geared toward specific cellular processes, such as Th17 immune response and morphology of macrophages (Figure 4C). The one cancer-related target of hsa-mir-3648 is CCND1 (G1/Sspecific cyclin-D1) involved in cell cycle control.
Table 4.
Newly identified SDEmiRNAs and the potential networks and pathways involved in cancer
| SDEmiRNA | Number of non-tissue-specific target | Networks and Pathways |
|---|---|---|
| hsa-mir-1269a | 57 | Molecular transport, RNA trafficking, cell death and survival Lipid metabolism, nucleic acid metabolism Cellular assembly and organization, DNA replication, recombination, and repair, cell morphology Cardiovascular system development and function, organismal development |
| hsa-mir-4652 | 133 | Cellular development, cellular growth and proliferation Organ Morphology, organismal development, organismal injury and abnormalities Cardiovascular system development and function, skeletal and muscular system development and function Immune response |
| hsa-mir-4746 | 54 | Skeletal and muscular disorders, organismal development Cellular assembly and organization, nervous system development and function, tissue morphology Cellular development, cellular growth and proliferation Intracellular membrane-bounded organelle |
| hsa-mir-3648 | 14 | Connective tissue disorders, development disorder, endocrine system disorders |
| hsa-mir-3687 | 14 | Cellular development, cellular growth and proliferation, cell cycle |
Figure 4. Networks and pathways of non-tissue-specific cancer-related targets of hsa-mir-1269a, hsa-mir-4652, and hsa-mir-4746.
A. Four out of five non-tissue-specific cancer-related targets of hsa-mir-1269a with grey shapes participate in 15 cellular processes with white shapes. Each arrow represents one correlation between a target and a process. GPI, TCF12, and SCD are involved in most of the processes, compared to PLAG1. B. Network of eight out of 12 cancer-related targets of hsa-mir-4652. These targets all participate in multiple cellular processes. C. Network of all six cancer-related targets of hsa-mir-4746. Except BLVRB, other targets share similar processes with each other. Based on the important roles of these cellular processes in cancer development, the three newly identified SDEmiRNAs may function in cancers through these processes.
By targeting a wide range of cancer-related genes, 65 SDEmiRNAs, especially 21 key SDEmiRNAs, participate in multiple processes, contributing to cancer development. Moreover, those newly identified SDEmiRNAs could be equally important as other well-studied miRNAs, despite their relatively lower expression levels. This study suggests that SDEmiRNAs may contribute to cancer by regulating targets within small regions or participating in short-term stress response, which requires less dosage.
Distribution and functional analysis of tissue-specific targets of 65 SDEmiRNAs
We also collected tissue-specific targets for the 65 SDEmiRNAs (significantly expressed in at least eight cancer types) (Table S3) and explored the potential molecular mechanisms of those tissue-specific targets in light of their observed expression trends and biological processes in which they participate. All selected miRNAs and their targets were mapped to 11 cancer types by DOIDs. A total of 9,572 target genes were found, of which 7,325 were mapped to reviewed UniProtKB accession numbers (Table S4). 7,186 of the implicated significant, reviewed target genes are included in BioXpress v3.0, which reports genes with significant expression changes derived from paired tumor and non-tumorous data from TCGA: currently, there are 5,971 genes showing significant differential expression in multiple cancer types in BioXpress (Table S5). miRNA target genes for lung cancer and kidney cancer are a combination of those identified for TCGA cancer types LUAD and LUSC, KICH and KIRP and KIRC, respectively, and have the same expression trend across these TCGA cancer types.
Mapping to Gene Ontology (GO) terms and PANTHER pathway IDs resulted in retrieval of 12,467 GO terms and 130 PANTHER terms. According to the screening conditions described in the methods section, 99 terms were selected in Table S6 (22 PANTHER terms and 77 GO terms). The criteria of the screening were such that we expected to lose potentially valuable GO or PANTHER terms, but we chose to follow a conservative method to reduce possible false positive results. The most common molecular functions in which these miRNAs and their targets participate in cancer are negative regulation of ERK1 and ERK2 cascade (GO:0070373), canonical Wnt signaling pathway (GO:0060070), and positive regulation of sequence-specific DNA binding transcription factor activity (GO:0051091). Each of these functions are involved in ten types of cancer, with more than 40 SDEmiRNAs, and more than 30 targets, respectively. This result not only is consistent with Gosline et al. [48], which suggested the strong regulation of miRNAs on transcription factors in mice, but also suggests most of these SDEmiRNAs could promote the activity of transcription factors.
The distribution of related GO and PANTHER terms suggests the participation in regulation of cell proliferation and apoptosis through different mechanisms, and frequently implicated cancer pathways were found in our list, including the p53 pathway, Wnt signaling pathway, TGFβ/SMAD signaling pathway, and others. We also found that the occurrence and development of these cancer types may share similar molecular mechanisms with other diseases or processes, including Alzheimer’s disease (AD) (with involved amyloid secretase and presenilin pathways), for which the product the key gene implicated in AD, the amyloid precursor protein, is overexpressed in multiple cancer types [49] and other neurodegenerative diseases.
Of the 21 key SDEmiRNAs (with both significant differential expression and at least 60% patient frequencies in at least eight cancer types) (Table 3), in addition to hsa-mir-4652, 19 were found to have tissue-specific targets. We identified the top one or two genes that are targeted by the greatest number of SDEmiRNAs in each cancer type. In breast cancer, CCND1 and IGF1R (Insulin-like growth factor 1 receptor) are found to be the targets of nine and seven key SDEmiRNAs, respectively. IGF1R is also the most popular target of eight SDEmiRNAs in lung cancer, and of seven SDEmiRNAs in head and neck cancer. CCND1 promotes G1/S transition during mitotic cell cycle by regulating CDK4 (cyclin-dependent kinase 4), and is up-regulated in various cancer types including breast cancer [50, 51]. IGF1R is also a well-studied oncogene that allows cell proliferation and growth, and inhibits apoptosis [52]. Our RNA-seq analysis in BioXpress v3.0 shows that CCND1 is significantly over-expressed in BRCA, while IGF1R has significant over-expression in BRCA, LUSC, and HNSC. We suspect the roles of CCND1 and IGF1R could be vital for development of these cancers, compared to other cancer-related genes, based on the abundance of miRNAs that can regulate them.
Furthermore, in breast cancer, hsa-mir-1269a has one target (EN2, Homeobox protein engrailed-2) which is a sequence-specific DNA binding protein positively regulating transcription from RNA polymerase II promoter. hsa-mir-4652 has two targets (GALNT3, EIF4EBP1) in lung cancer. GALNT3 (Polypeptide N-acetylgalactosaminyltransferase 3) functions as an iron binding enzyme in glycosylation, while EIF4EBP1 (Eukaryptic translation initiation factor 4E-binding protein 1) binds to IRES-dependent translational initiation to repress protein translation and participates in insulin receptor signaling pathway and G1/S transition of mitotic cell cycle (Table 4). hsa-mir-4652 has another target (H2AFX, Histone H2AX) involved in DNA and histone binding in head and neck cancer. CancerMiner predicted 12 targets of hsa-mir-3648 in head and neck cancer (Table S4) with functions including ATP binding, cell cycle control, lipid metabolism, protein folding, and nucleobase biosynthesis. hsa-mir-4746 has a common target, PPM1D (Protein phosphatase 1D), in both breast and lung cancer. Because of its function in protein dephosphorylation, PPM1D is involved in G2/M transition of mitotic cell cycle and suppression of cell proliferation in cancer [53]. The importance of the functions of their target genes in gene/cellular regulation suggests abnormally expressed hsa-mir-1269a, hsa-mir-4652, and hsamir-4746 may contribute to the development of corresponding cancer types.
Based on their expression profiles in at least eight cancer types, as well as the cellular functions of their cancer-related targets, those SDEmiRNAs with conserved expression change trend in at least eight cancers, especially the 21 key SDEmiRNAs with high patient frequencies, show great potential as drivers in the development of multiple cancers. Our study first identified the common essential miRNAs across different cancers, followed by identification of potential common molecular functions despite different tissues. This list should guide the direction of future cancer research efforts and increase the efficiency of discovering detailed mechanisms across cancers by targeting the scope of study to high-value miRNA candidates.
Construction of regulatory networks of SDEmiRNAs and their targets
Suppression of a negative regulator will result in up-regulation of the regulator’s target molecule. Similarly, because miRNAs function to suppress gene expression, miRNA inhibition of a negative regulator will also result in up-regulation of the target molecule. There are some cases in which a miRNA can have two targets such that one target regulates the other. In these cases, up-regulated targets of over-expressed miRNAs or down-regulated targets of under-expressed miRNAs are considered to be indirect targets: miRNAs either inhibit the negative regulators or activate the positive regulators of the differentially expressed target genes, respectively.
Across eight cancer types, there are 34 SDEmiRNAs that regulate 14 potential target genes in the angiogenic patterning of blood vessels (GO:0001569), which is essential for establishing abnormal tumor blood vessels [54]. Our results here show hsa-mir106b, hsa-mir-93, and hsamir-21, which are over-expressed in six cancers and target eight genes (out of the 14 possible targets), play important roles in tumor blood vessel generation. All three SDEmiRNAs directly target one key tumor suppressor gene, TGFBR2 (TGF-beta receptor II) [55], leading to its under-expression. Unlike over-expressed miRNAs with a relatively large number of targets, most under-expressed miRNAs only target central genes of angiogenesis, including TGFBR2 (indirect), VEGFA (vascular endothelial growth factor A) (direct), and CTNNB1 (beta-catenin) (direct). These trends could suggest that during patterning of blood vessels in these eight cancer types, most tumor suppressive miRNAs regulating angiogenic central genes are repressed, while the three oncogenic miRNAs above activate a variety of relevant genes, efficiently promoting the tumorigenic process.
To further elucidate the underlying mechanisms of miRNA involvement in cancer, we constructed a network of miRNA regulation of signaling pathways across different cancer types. The PI3-kinase (PI3K) pathway (PANTHER: P00048) is one of the most important pathways that participates in the regulation of cancer development and growth [56]. PTEN, an important tumor suppressor, is a negative regulator of the PI3K pathway [57]. As shown in Figure 5, eight target genes of 16 miRNAs are involved in the PI3K pathway across seven cancer types, based on KEGG (Release 79.1, http://www.genome.jp/kegg/) and studies from Waugh MG [58], Ying Z et al. [59], and Yun YR et al. [60]. miRNAs involved in regulating FGFR family members were also shown in Figure 5, adding another potentially important miRNA, hsa-mir-381, to the pathway.
Figure 5. Regulatory network of miRNA on PI3-kinase pathway.
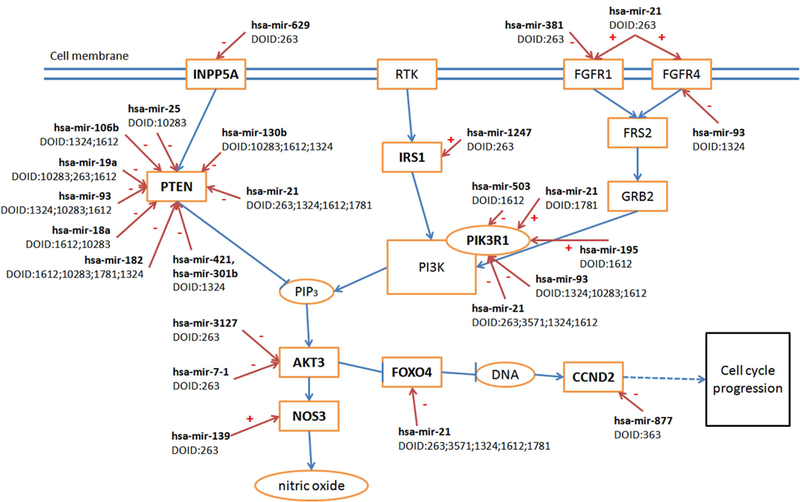
Blue arrows indicate the regulation among genes in the PI3-kinase pathway (arrows with vertical short line segments represent inhibition role of up-stream genes). Red arrows indicate regulation interaction between miRNAs and PI3-kinase pathway. Minus sign suggests inhibition, and plus sign promotion. Eight target genes of 16 miRNAs are involved in the PI3K pathway and regulation of PTEN gene across seven cancer types. Different miRNAs in different cancers could regulate the same molecular processes through targeting their components. These miRNAs still could function similarly across at least eight cancer types.
Most over-expressed miRNAs silence PTEN across different cancer types, increasing the signal passing from PIK3 to AKT3, and some miRNAs regulate PI3KR1 (phosphoinositide-3kinase regulatory subunit 1, a gene of PI3K Class I). hsa-mir-21, for instance, is a well-known, over-expressed miRNA in various cancer types. Our results in Figure 5 indicate that hsa-mir-21 could inhibit PI3KR1 in breast, liver, kidney, and lung cancer, while it promotes PI3KR1 in thyroid cancer, suggesting that for cancer types other than thyroid cancer, other members of the PI3K classes of enzymes may play more important roles in cancer progression. Furthermore, we suggest that different SDEmiRNAs could still regulate the same molecular processes in different cancer types, even if their target genes may vary from each other. This network integrates current discoveries on SDEmiRNAs, their targets, and related functions, displays SDEmiRNA regulation in multiple cancer types, and, therefore, provides a straightforward view of what could be accomplished in additional analysis. Similar networks could be constructed based on other results reported herein, and could ultimately lay the foundation for future targeted studies regarding the role of miRNA expression in cancer.
Materials and Methods
Data
miRNA sequencing data
This study started with miRNA sequencing data integration and filtering, and performed expression analysis and downstream functional analysis (Figure 6). All miRNA-seq data (both counts of raw reads and Reads Per Million miRNA mapped (RPM) values, annotated to miRNA names level) were downloaded from TCGA (Release-2016–03) (http://cancergenome.nih.gov) by using TCGA-Assembler [61] with default parameters (Figure 6). Data was then divided into two groups: paired data—data with both tumor and adjacent non-tumorous samples from the same patient, and tumor-only expression data—data coming exclusively from all tumor samples in all TCGA cancer types with miRNA-seq data. Paired data were limited to those generated on the HiSeq platform which generated more matched samples than other platforms, whereas tumor-only expression data were focused on data generated either on HiSeq or Genome Analyzer (GA) of Illumina [62] systems, analyzed in separate groups for data from each platform. Two additional projects (MALY, Malignant Lymphoma done by Germany, and OV, Ovarian Cancer done by Australia, Release 20), available only through the International Cancer Genome Consortium (ICGC, https://icgc.org/) and generated by Illumina-HiSeq system, were imported into the tumor-only expression dataset. Fold change value (FC) was calculated by (RPM)tumor / (RPM)non-tumor for one miRNA in one cancer type. Each miRNA was mapped to its corresponding HGNC ID (HUGO Gene Nomenclature Committee, Issued in Autumn/Winter 2015: http://www.genenames.org), miRBase ID (Release 21) [1], and Ensembl ID (Release 82) [63] using customized python (v2.7) and R (3.3) scripts.
Figure 6. Cancer-related significantly differential expression of miRNA workflow.
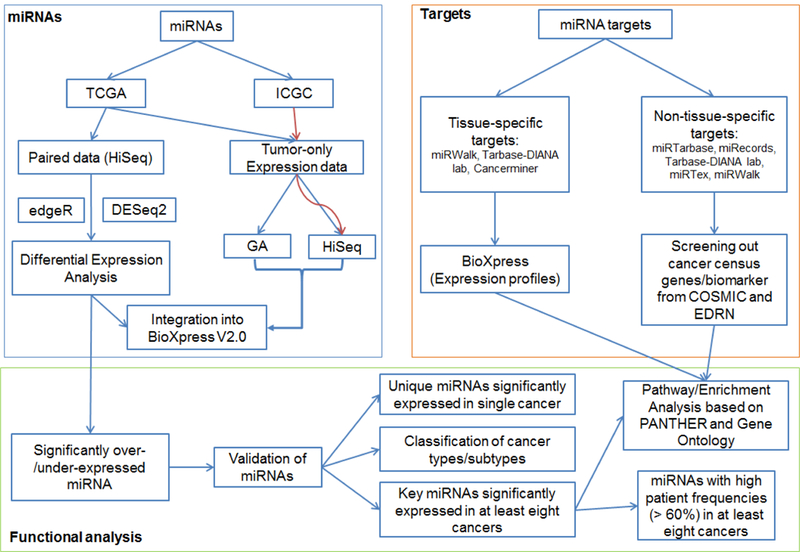
Paired miRNAs (tumorous and adjacent non-tumorous samples) were retrieved from TCGA database and analyzed by DESeq2 and edgeR for differential expression. We then screened out those significantly differentially expressed miRNAs (SDEmiRNAs). After validation by text mining databases and our text mining tool, DEXTER, we proposed the possibility to use these SDEmiRNAs in classification of cancer types or cancer subtypes, and screened out unique SDEmiRNAs for one single cancer. We also kept those key SDEmiRNAs significantly expressed in at least eight cancers with high patient frequencies. Moreover, we collected both tissuespecific targets and non-tissue-specific targets from experimentally validated databases, and did functional/enrichment analysis with those key SDEmiRNAs.
Databases for Validation of miRNA
To validate the selected differentially expressed miRNAs, we used four experimentally validated databases generated from text mining: HMDD (v2.0) [64], Mir2disease (2008) [65], miRCancer (March2016) [66], and miRiaD (2016) [67]. We also applied our text mining tool, DEXTER, to extract miRNA-cancer correlations from publications [41]. The basic search term is “ [miRNA name] is [over-expressed / under-expressed / significantly higher / lower] in [cancer tissue/sample] compared to [normal / non-tumorous sample]”. Results were manually curated before reported as validated records. The cancer terms for each reported interaction between miRNA and specific cancer types were then mapped to Disease Ontology [68] cancer slim in order to unify the integrated dataset [69]. We also incorporated information regarding miRNA expression changes in cancer types (up/down-regulation) from these databases, when available.
miRNA Targets
Five experimentally validated miRNA target databases generated from text mining were used in our study: miRTarbase (Release 6.0) [70], miRecords (Apr 2013) [71], miRWalk 2.0 (miRNAtarget interaction databases) [72], miRTex (May 2015) [73], and Tarbase-DIANA lab v6.0 [74]. These databases were generated by different algorithms but with a common motive, which is to extract miRNA-target-cancer/tissue interactions from published papers. Most of these published papers were validating the interactions or stating that the interactions were commonly accepted by previous studies. CancerMiner (1.0), a database for tissue-specific miRNA target predictions based on TCGA data [75], was also included in the integrated miRNA target database for this study. This database was built upon multivariate linear model for expression association of miRNA and mRNA from TCGA, and rank-based association recurrence score that showed the possible association of miRNA-target interactions and cancer types [75].
GO (Gene Ontology) IDs [76] and PANTHER pathway IDs and terms were downloaded from UniProt (http://www.uniprot.org/, Release 2016_08) and PANTHER ((Protein ANalysis THrough Evolutionary Relationships, Release 11.0: http://www.pantherdb.org/) [77] for each target in the resulting database.
Data Normalization and Differential Expression Analysis
We used DESeq2 version 1.8.1 [28] and edgeR version 3.10.2 [26] (R packages) to normalize paired miRNA sequencing data and to analyze the differential expression of miRNAs in each cancer type. These tools are both based on a negative binomial distribution assumption [29, 39], and both tools have been proven to generate normalized data of high quality and to perform similarly in differential expression analyses, as well as in fold change estimates [78, 79]. DESeq2 fits a generalized linear model for each miRNA with the fold change estimate shrunken by empirical Bayes [28]. A Wald test was used in DESeq2 for statistical analysis and calculating p values for the significance of differentially expressed miRNAs in each cancer type. edgeR, however, uses the TMM method for normalization of sequencing data. To estimate parameters for the negative binomial model, we applied common dispersion, trended dispersion, and tagwise dispersion for our multi-factor cases. These dispersions were shrunken by an empirical Bayes procedure towards a consensus value [26], and edgeR applied Fisher’s Exact Test to determine differential expression [26]. False discovery rate (FDR) was reported by edgeR for the significance of multiple comparisons conducted. The input data for the two tools were raw read counts (without normalization), excluding any miRNAs with no expression among all patients. For both tools, we designed the model matrix with two categories, one with levels “cancer” and “non-tumor”, the other with TCGA patient IDs. The cutoff for the significant adjusted p-value of each gene is 0.05/n (using Bonferroni’s Approach, where n is the total number of expressed miRNAs in each cancer type), considering the difference of cancers. miRNAs with significant adjusted p-values generated by both DESeq2 and edgeR were considered to be differentially expressed in a given cancer type. Results from differential expression analysis for paired and tumor-only expression data were integrated into BioXpress version 3.0 [80]. The Heatmap with clustering for significantly differentially expressed miRNAs across different cancer types was generated by R package gplots 3.0.1.
Survival Analysis
miRpower [40] was used to suggest the influence of miRNAs on patient survival rate in breast cancer. This platform is continuously updated with TCGA datasets, and has been cited by dozens of studies [81, 82, 83]. By providing “auto select best cutoff”, miRpower is able to optimize the cutoff of the patient groups for different investigated miRNAs. In our study here, TCGA was selected as the only dataset to be considered, and therefore, miRNAs from breast cancer (BRCA) were under survival analysis (1,061 TCGA patients in total).
miRNA Target Data Integration and Enrichment
After extracting all targets for selected miRNAs (those significantly over- or under-expressed in at least eight cancer types), we separated these targets into two categories: tissue-specific targets (records clearly including the miRNA-target connection in a specific tissue or cancer type), and non-tissue-specific targets (records containing only a miRNA-target connection). Tarbase provided the majority of records for the tissue-specific target group. We excluded associations derived from microarray studies and integrated those records with cancer metastasis annotation. Reported regulatory changes (up/down) of miRNAs were also retrieved. Selective records from miRWalk 2.0 were included in the tissue-specific target group if the records were involved in miRNAs-organ interactions or miRNA-gene-OMIM interactions generated by miRWalk text mining search on PubMed database. To include more tissue-specific targets, we also integrated data from a predicted database, CancerMiner, which applied mRNA and miRNA expression, DNA copy number, and promoter methylation data from TCGA to predict miRNA-mRNA-cancer type associations [75]. CancerMiner data were included only for records not found in Tarbase or miRWalk. Expression changes of all tissue-specific target genes in corresponding cancer types were extracted from BioXpress v3.0 [80]. For the non-tissuespecific target group, we first excluded those targets that were also in the tissue-specific target section. Some records from Tarbase that do not contain tissue or cancer information were included. Gene names of the records from miRTex were first manually curated and mapped to synonyms available in UniProtKB, and records with miRNA-gene associations were selected while miRNA-miRNA interactions were excluded. miRNA-gene targets from miRWalk 2.0 and records from miRTarbase and miRecords were all classified into the non-tissue-specific target group. All cancer types from those databases were mapped to DOIDs.
Tissue-specific target genes were mapped to UniProtKB accession numbers [84], maintaining only those genes that have been manually annotated and reviewed in UniProtKB/Swiss-Prot. For each GO or PANTHER term, we merged all the target genes involved and their regulating miRNAs. In order to reduce false positive errors and to exclude highly generic terms of GO, GO terms of genes associated more than eight cancer types and fewer than 50 target genes were selected. We also collected non-tissue-specific targets that are considered to be cancer census genes from COSMIC (version 78) [85] and biomarkers from EDRN (Downloaded in Feb, 2016, https://edrn.nci.nih.gov/). Enrichment analysis of these non-tissue-specific targets was done using STRING (version 10.0) [86] and Ingenuity Pathway Analysis (IPA) (March 2017).
Conclusion
Different cancer types have unique expression patterns of SDEmiRNAs. Our study sought to identify prevalent expression-based or mechanistic features of SDEmiRNAs associated with cancer development.. Applying our miRNA analysis findings, we propose that the number of miRNAs needed to classify the 14 cancer types could be reduced, both in the cancers described herein and in other cell type cancers from the same location. Moreover, based on the inhibition role of miRNA, development of different cancer types may rely on either more tumor suppressors or more oncogenes, resulting in three categories of cancers. We also identified 81 unique SDEmiRNAs with significant differential expression in specific cancers: these SDEmiRNAs might have unique functions (including potential roles as drivers) in each cancer, even affecting patient survival rates, although most have not yet been studied in the corresponding relevant cancer type.
We then identified a set of 21 key SDEmiRNAs (including four relatively recent identifications, hsa-mir-4746, hsa-mir-3648, hsa-mir-3687, and hsa-mir-1269a) that have high enrichment in patients for at least eight cancer types, as well as hsa-mir-4652 with high enrichment in seven cancer types. These key SDEmiRNAs may have a wider scope of potential regulatory mechanisms across multiple cancer types while acting similarly despite different cancer types than what researchers have reported in one or two cancers so far. To explore the possible molecular mechanisms of these key SDEmiRNAs, enrichment analysis of the SDEmiRNAs and their targets in different cancer types was performed, findings of which suggested that different miRNAs participate in a limited amount of cancer-associated molecular processes/pathways through regulating different targets. To the best of our knowledge, this is the first report suggesting that miRNAs may use similar mechanisms in development of these cancer types.
The construction of a regulatory network of SDEmiRNAs with their experimentally validated targets combines results of current studies and highlights the potential research directions for further analysis. Our study here provides a rationale for the continued exploration and validation of the functional roles of selected key SDEmiRNAs in cancers, and, therefore, promotes on the use of BioXpress to facilitate further analysis on the role of these important miRNAs in cancer.
Supplementary Material
Highlights.
A large-scale investigation of miRNAs between tumor and adjacent nontumorous samples
A novel perspective to group cancer types based on the expression profiles of miRNAs
Identification of common potential biomarker miRNAs across eight or more cancer types
A method to build common regulatory networks of key miRNAs across multiple cancers
Acknowledgments
The authors thank Yu Fan and Ting-Chia Chang for helpful discussion. Yu Hu earned an MS in the Bioinformatics Track of the Bioinformatics and Molecular Biochemistry Program at the George Washington University.
Funding
This project was partially funded by NCI/EDRN Associate Member contract #156620 and The McCormick Genomic and Proteomic Center (MGPC), the George Washington University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interest
None.
Additional Information
Results of miRNA differential expression analysis are available at: https://hive.biochemistry.gwu.edu/bioxpress
References
- [1].Kozomara A, Griffiths-Jones S, miRBase: annotating high confidence microRNAs using deep sequencing data, Nucleic acids research, 42 (2014) D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zempleni J, Baier SR, Howard KM, Cui J, Gene regulation by dietary microRNAs, Canadian journal of physiology and pharmacology, 93 (2015) 1097–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM, Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia, Proceedings of the National Academy of Sciences of the United States of America, 99 (2002) 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T, Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival, Cancer research, 64 (2004) 3753–3756. [DOI] [PubMed] [Google Scholar]
- [5].Chan JA, Krichevsky AM, Kosik KS, MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells, Cancer research, 65 (2005) 6029–6033. [DOI] [PubMed] [Google Scholar]
- [6].Ma L, Teruya-Feldstein J, Weinberg RA, Tumour invasion and metastasis initiated by microRNA-10b in breast cancer, Nature, 449 (2007) 682–688. [DOI] [PubMed] [Google Scholar]
- [7].Gonzalez-Quintana V, Palma-Berre L, Campos-Parra AD, Lopez-Urrutia E, Peralta-Zaragoza O, Vazquez-Romo R, Perez-Plasencia C, MicroRNAs are involved in cervical cancer development, progression, clinical outcome and improvement treatment response (Review), Oncology reports, 35 (2016) 3–12. [DOI] [PubMed] [Google Scholar]
- [8].Tutar L, Tutar E, Ozgur A, Tutar Y, Therapeutic Targeting of microRNAs in Cancer: Future Perspectives, Drug development research, 76 (2015) 382–388. [DOI] [PubMed] [Google Scholar]
- [9].Wang W, Luo YP, MicroRNAs in breast cancer: oncogene and tumor suppressors with clinical potential, Journal of Zhejiang University. Science. B, 16 (2015) 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hayes J, Peruzzi PP, Lawler S, MicroRNAs in cancer: biomarkers, functions and therapy, Trends in molecular medicine, 20 (2014) 460–469. [DOI] [PubMed] [Google Scholar]
- [11].Thomas J, Ohtsuka M, Pichler M, Ling H, MicroRNAs: Clinical Relevance in Colorectal Cancer, International journal of molecular sciences, 16 (2015) 28063–28076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Khan MA, Zubair H, Srivastava SK, Singh S, Singh AP, Insights into the Role of microRNAs in Pancreatic Cancer Pathogenesis: Potential for Diagnosis, Prognosis, and Therapy, Advances in experimental medicine and biology, 889 (2015) 71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zeng X, Zhang X, Zou Q, Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks, Briefings in bioinformatics, 17 (2016) 193–203. [DOI] [PubMed] [Google Scholar]
- [14].Zou Q, Li J, Song L, Zeng X, Wang G, Similarity computation strategies in the microRNAdisease network: a survey, Briefings in functional genomics, 15 (2016) 55–64. [DOI] [PubMed] [Google Scholar]
- [15].Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR, MicroRNA expression profiles classify human cancers, Nature, 435 (2005) 834–838. [DOI] [PubMed] [Google Scholar]
- [16].Ren Z, Wang W, Li J, Identifying molecular subtypes in human colon cancer using gene expression and DNA methylation microarray data, International journal of oncology, 48 (2016) 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sisti JS, Collins LC, Beck AH, Tamimi RM, Rosner BA, Eliassen AH, Reproductive risk factors in relation to molecular subtypes of breast cancer: Results from the nurses’ health studies, International journal of cancer, 138 (2016) 2346–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gasparini P, Cascione L, Landi L, Carasi S, Lovat F, Tibaldi C, Ali G, D’Incecco A, Minuti G, Chella A, Fontanini G, Fassan M, Cappuzzo F, Croce CM, microRNA classifiers are powerful diagnostic/prognostic tools in ALK-, EGFR-, and KRAS-driven lung cancers, Proceedings of the National Academy of Sciences of the United States of America, 112 (2015) 14924–14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Monroig-Bosque Pdel C, Rivera CA, Calin GA, MicroRNAs in cancer therapeutics: “from the bench to the bedside”, Expert opinion on biological therapy, 15 (2015) 1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sethi S, Ali S, Sarkar FH, MicroRNAs in personalized cancer therapy, Clinical genetics, 86 (2014) 68–73. [DOI] [PubMed] [Google Scholar]
- [21].Peng F, Zhang Y, Wang R, Zhou W, Zhao Z, Liang H, Qi L, Zhao W, Wang H, Wang C, Guo Z, Gu Y, Identification of differentially expressed miRNAs in individual breast cancer patient and application in personalized medicine, Oncogenesis, 5 (2016) e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Metpally RP, Nasser S, Malenica I, Courtright A, Carlson E, Ghaffari L, Villa S, Tembe W, Van Keuren-Jensen K, Comparison of Analysis Tools for miRNA High Throughput Sequencing Using Nerve Crush as a Model, Frontiers in genetics, 4 (2013) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Humphreys DT, Suter CM, miRspring: a compact standalone research tool for analyzing miRNA-seq data, Nucleic acids research, 41 (2013) e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lopes-Ramos CM, Habr-Gama A, Quevedo Bde S, Felicio NM, Bettoni F, Koyama FC, Asprino PF, Galante PA, Gama-Rodrigues J, Camargo AA, Perez RO, Parmigiani RB, Overexpression of miR-21–5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients, BMC medical genomics, 7 (2014) 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cobos Jimenez V, Willemsen AM, Bradley EJ, Baas F, van Kampen AH, Kootstra NA, Nextgeneration sequencing of microRNAs in primary human polarized macrophages, Genomics data, 2 (2014) 181–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 26 (2010) 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Anders S, Huber W, Differential expression analysis for sequence count data, Genome biology, 11 (2010) R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNAseq data with DESeq2, Genome biology, 15 (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yang S, Guo L, Shao F, Zhao Y, Chen F, A Systematic Evaluation of Feature Selection and Classification Algorithms Using Simulated and Real miRNA Sequencing Data, Computational and mathematical methods in medicine, 2015 (2015) 178572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Funamizu N, Lacy CR, Parpart ST, Takai A, Hiyoshi Y, Yanaga K, MicroRNA-301b promotes cell invasiveness through targeting TP63 in pancreatic carcinoma cells, International journal of oncology, 44 (2014) 725–734. [DOI] [PubMed] [Google Scholar]
- [31].Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH, Chang YC, Lin WC, Shen CY, Huang CS, Hsieh FJ, Lai LC, Tsai MH, Chang KJ, Chuang EY, Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing, Molecular cancer, 14 (2015) 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tang D, Zhang Q, Zhao S, Wang J, Lu K, Song Y, Zhao L, Kang X, Xu S, Tian L, The expression and clinical significance of microRNA-1258 and heparanase in human breast cancer, Clinical biochemistry, 46 (2013) 926–932. [DOI] [PubMed] [Google Scholar]
- [33].Liu H, Chen X, Gao W, Jiang G, The expression of heparanase and microRNA-1258 in human non-small cell lung cancer, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 33 (2012) 1327–1334. [DOI] [PubMed] [Google Scholar]
- [34].Bu P, Wang L, Chen KY, Rakhilin N, Sun J, Closa A, Tung KL, King S, Kristine Varanko A, Xu Y, Huan Chen J, Zessin AS, Shealy J, Cummings B, Hsu D, Lipkin SM, Moreno V, Gumus ZH, Shen X, miR-1269 promotes metastasis and forms a positive feedback loop with TGF-beta, Nature communications, 6 (2015) 6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gonul O, Aydin HH, Kalmis E, Kayalar H, Ozkaya AB, Atay S, Ak H, Effects of Ganoderma lucidum (Higher Basidiomycetes) Extracts on the miRNA Profile and Telomerase Activity of the MCF-7 Breast Cancer Cell Line, International journal of medicinal mushrooms, 17 (2015) 231–239 [DOI] [PubMed] [Google Scholar]
- [36].Emmadi R, Canestrari E, Arbieva ZH, Mu W, Dai Y, Frasor J, Wiley E, Correlative Analysis of miRNA Expression and Oncotype Dx Recurrence Score in Estrogen Receptor Positive Breast Carcinomas, PloS one, 10 (2015) e0145346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, Loman N, Jonsson G, Naya H, Hoglund M, Borg A, Rovira C, Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene, Cancer research, 71 (2011) 78–86 [DOI] [PubMed] [Google Scholar]
- [38].Tang W, Wan S, Yang Z, Teschendorff AE, Zou Q, Tumor origin detection with tissue-specific miRNA and DNA methylation markers, Bioinformatics, 34 (2018) 398–406. [DOI] [PubMed] [Google Scholar]
- [39].Varet H, Brillet-Gueguen L, Coppee JY, Dillies MA, SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data, PloS one, 11 (2016) e0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, Gyorffy B, miRpower: a webtool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients, Breast cancer research and treatment, 160 (2016) 439–446. [DOI] [PubMed] [Google Scholar]
- [41].Gupta S, Dingerdissen H, Ross KE, Hu Y, Wu CH, Mazumder R, Vijay-Shanker K, DEXTER: Disease-Expression Relation Extraction from Text, Database : the journal of biological databases and curation, 2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu Y, Zeng X, He Z, Zou Q, Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources, IEEE/ACM transactions on computational biology and bioinformatics, 14 (2017) 905–915. [DOI] [PubMed] [Google Scholar]
- [43].Zou Q, Li J, Hong Q, Lin Z, Wu Y, Shi H, Ju Y, Prediction of MicroRNA-Disease Associations Based on Social Network Analysis Methods, BioMed research international, 2015 (2015) 810514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ, A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome, Annual review of genetics, 49 (2015) 213–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Min P, Li W, Zeng D, Ma Y, Xu D, Zheng W, Tang F, Chen J, Shi J, Hu H, Wang J, Yang D, Liu J, Zhang J, Zhang M, A single nucleotide variant in microRNA-1269a promotes the occurrence and process of hepatocellular carcinoma by targeting to oncogenes SPATS2L and LRP6, Bulletin du cancer, 104 (2017) 311–320. [DOI] [PubMed] [Google Scholar]
- [46].Ma Y, Liang AJ, Fan YP, Huang YR, Zhao XM, Sun Y, Chen XF, Dysregulation and functional roles of miR-183-96-182 cluster in cancer cell proliferation, invasion and metastasis, Oncotarget, (2016). [DOI] [PMC free article] [PubMed]
- [47].Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR, A census of human cancer genes, Nature reviews. Cancer, 4 (2004) 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gosline SJ, Gurtan AM, JnBaptiste CK, Bosson A, Milani P, Dalin S, Matthews BJ, Yap YS, Sharp PA, Fraenkel E, Elucidating MicroRNA Regulatory Networks Using Transcriptional, Posttranscriptional, and Histone Modification Measurements, Cell reports, 14 (2016) 310–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Woods NK, Padmanabhan J, Inhibition of amyloid precursor protein processing enhances gemcitabine-mediated cytotoxicity in pancreatic cancer cells, The Journal of biological chemistry, 288 (2013) 30114–30124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Long J, Ou C, Xia H, Zhu Y, Liu D, MiR-503 inhibited cell proliferation of human breast cancer cells by suppressing CCND1 expression, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 36 (2015) 8697–8702. [DOI] [PubMed] [Google Scholar]
- [51].Reis-Filho JS, Savage K, Lambros MB, James M, Steele D, Jones RL, Dowsett M, Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis, Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc, 19 (2006) 999–1009. [DOI] [PubMed] [Google Scholar]
- [52].Werner H, Sarfstein R, Transcriptional and epigenetic control of IGF1R gene expression: implications in metabolism and cancer, Growth hormone & IGF research : official journal of the Growth Hormone Research Society and the International IGF Research Society, 24 (2014) 112–118. [DOI] [PubMed] [Google Scholar]
- [53].Ogasawara S, Kiyota Y, Chuman Y, Kowata A, Yoshimura F, Tanino K, Kamada R, Sakaguchi K, Novel inhibitors targeting PPM1D phosphatase potently suppress cancer cell proliferation, Bioorganic & medicinal chemistry, 23 (2015) 6246–6249. [DOI] [PubMed] [Google Scholar]
- [54].Nagy JA, Chang SH, Dvorak AM, Dvorak HF, Why are tumour blood vessels abnormal and why is it important to know?, British journal of cancer, 100 (2009) 865–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Xu JB, Bao Y, Liu X, Liu Y, Huang S, Wang JC, Defective expression of transforming growth factor beta type II receptor (TGFBR2) in the large cell variant of non-small cell lung carcinoma, Lung Cancer, 58 (2007) 36–43. [DOI] [PubMed] [Google Scholar]
- [56].Baselga J, Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer, The oncologist, 16 Suppl 1 (2011) 12–19. [DOI] [PubMed] [Google Scholar]
- [57].Chalhoub N, Baker SJ, PTEN and the PI3-kinase pathway in cancer, Annual review of pathology, 4 (2009) 127–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Waugh MG, Chromosomal Instability and Phosphoinositide Pathway Gene Signatures in Glioblastoma Multiforme, Molecular neurobiology, 53 (2016) 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ying Z, Xie X, Chen M, Yi K, Rajagopalan S, Alpha-lipoic acid activates eNOS through activation of PI3-kinase/Akt signaling pathway, Vascular pharmacology, 64 (2015) 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yun YR, Won JE, Jeon E, Lee S, Kang W, Jo H, Jang JH, Shin US, Kim HW, Fibroblast growth factors: biology, function, and application for tissue regeneration, Journal of tissue engineering, 2010 (2010) 218142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhu Y, Qiu P, Ji Y, TCGA-assembler: open-source software for retrieving and processing TCGA data, Nature methods, 11 (2014) 599–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Minoche AE, Dohm JC, Himmelbauer H, Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems, Genome biology, 12 (2011) R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Keenan S, Lavidas I, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Nuhn M, Parker A, Patricio M, Pignatelli M, Rahtz M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Birney E, Harrow J, Muffato M, Perry E, Ruffier M, Spudich G, Trevanion SJ, Cunningham F, Aken BL, Zerbino DR, Flicek P, Ensembl 2016, Nucleic acids research, 44 (2016) D710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, Cui Q, HMDD v2.0: a database for experimentally supported human microRNA and disease associations, Nucleic acids research, 42 (2014) D1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y, miR2Disease: a manually curated database for microRNA deregulation in human disease, Nucleic acids research, 37 (2009) D98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Xie B, Ding Q, Han H, Wu D, miRCancer: a microRNA-cancer association database constructed by text mining on literature, Bioinformatics, 29 (2013) 638–644. [DOI] [PubMed] [Google Scholar]
- [67].Hinske LC, Franca GS, Torres HA, Ohara DT, Lopes-Ramos CM, Heyn J, Reis LF, OhnoMachado L, Kreth S, Galante PA, miRIAD-integrating microRNA inter- and intragenic data, Database : the journal of biological databases and curation, 2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D, Parkinson H, Schriml LM, Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data, Nucleic acids research, 43 (2015) D1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wu TJ, Schriml LM, Chen QR, Colbert M, Crichton DJ, Finney R, Hu Y, Kibbe WA, Kincaid H, Meerzaman D, Mitraka E, Pan Y, Smith KM, Srivastava S, Ward S, Yan C, Mazumder R, Generating a focused view of disease ontology cancer terms for pan-cancer data integration and analysis, Database : the journal of biological databases and curation, 2015 (2015) bav032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY Tu SJ, Tsai TR, Ho SY, Jian TY, Wu HY, Chen PR, Lin NC, Huang HT, Yang TL, Pai CY, Tai CS, Chen WL, Huang CY, Liu CC, Weng SL, Liao KW, Hsu WL, Huang HD, miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database, Nucleic acids research, 44 (2016) D239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T, miRecords: an integrated resource for microRNAtarget interactions, Nucleic acids research, 37 (2009) D105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Dweep H, Gretz N, miRWalk2.0: a comprehensive atlas of microRNA-target interactions, Nature methods, 12 (2015) 69. [DOI] [PubMed] [Google Scholar]
- [73].Li G, Ross KE, Arighi CN, Peng Y, Wu CH, Vijay-Shanker K, miRTex: A Text Mining System for miRNA-Gene Relation Extraction, PLoS computational biology, 11 (2015) e1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG, TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support, Nucleic acids research, 40 (2012) D222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Jacobsen A, Silber J, Harinath G, Huse JT, Schultz N, Sander C, Analysis of microRNA-target interactions across diverse cancer types, Nature structural & molecular biology, 20 (2013) 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Consortium GO, Gene Ontology Consortium: going forward, Nucleic acids research, 43 (2015) D1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD, PANTHER version 10: expanded protein families and functions, and analysis tools, Nucleic acids research, 44 (2016) D336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloe D, Le Gall C, Schaeffer B, Le Crom S, Guedj M, Jaffrezic F, A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis, Briefings in bioinformatics, 14 (2013) 671–683. [DOI] [PubMed] [Google Scholar]
- [79].Tam S, Tsao MS, McPherson JD, Optimization of miRNA-seq data preprocessing, Briefings in bioinformatics, 16 (2015) 950–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Dingerdissen HM, Torcivia-Rodriguez J, Hu Y, Chang TC, Mazumder R, Kahsay R, BioMuta and BioXpress: mutation and expression knowledgebases for cancer biomarker discovery, Nucleic acids research, 46 (2018) D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhang K, Wang YW, Wang YY, Song Y, Zhu J, Si PC, Ma R, Identification of microRNA biomarkers in the blood of breast cancer patients based on microRNA profiling, Gene, 619 (2017) 10–20. [DOI] [PubMed] [Google Scholar]
- [82].Hawk MA, Gorsuch CL, Fagan P, Lee C, Kim SE, Hamann JC, Mason JA, Weigel KJ, Tsegaye MA, Shen L, Shuff S, Zuo J, Hu S, Jiang L, Chapman S, Leevy WM, DeBerardinis RJ, Overholtzer M, Schafer ZT, RIPK1-mediated induction of mitophagy compromises the viability of extracellular-matrix-detached cells, Nature cell biology, 20 (2018) 272–284. [DOI] [PubMed] [Google Scholar]
- [83].Xiao G, Zhang B, Meng J, Wang J, Xu C, Tang SC, Li X, Zhang J, Liang R, Ren H, Sun X, miR367 stimulates Wnt cascade activation through degrading FBXW7 in NSCLC stem cells, Cell cycle, 16 (2017) 2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Consortium U, UniProt: a hub for protein information, Nucleic acids research, 43 (2015) D204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA, COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer, Nucleic acids research, 39 (2011) D945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C, STRING v10: proteinprotein interaction networks, integrated over the tree of life, Nucleic acids research, 43 (2015) D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.