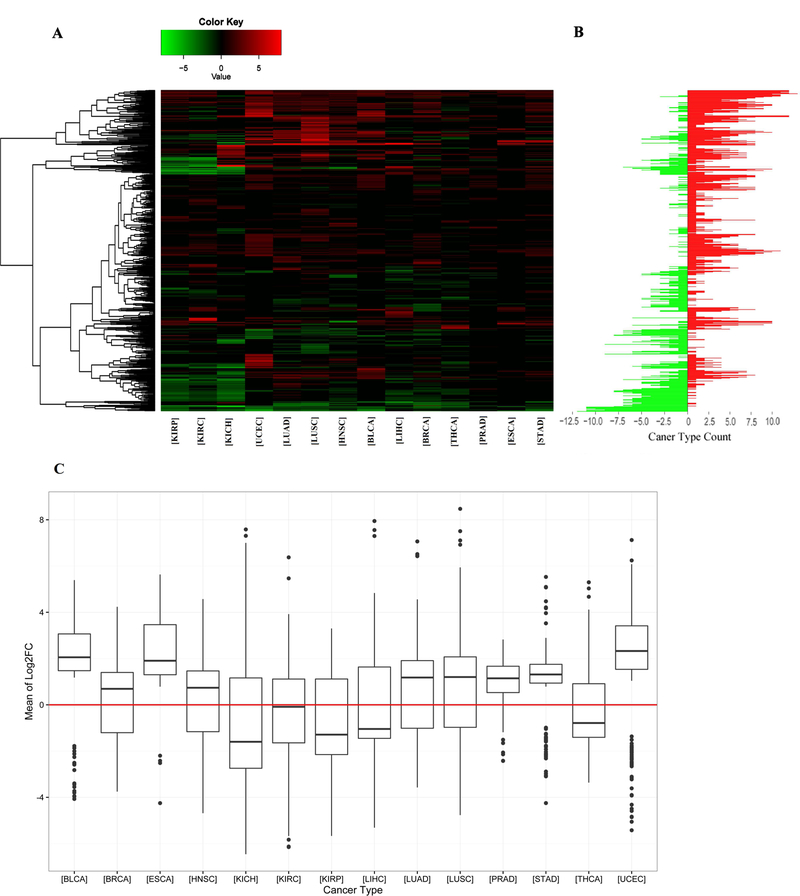
Figure 2. Differential expression analysis of miRNAs and distribution of log2FC values of significantly differentially expressed miRNAs across 14 cancer types.
A. Heatmap showing significantly over- and under-expressed miRNAs in at least one cancer type - vertical axis is miRNAs. The expression change of miRNA is represented by log2FC values generated by DESeq2 and edgeR. Log2FC values less than zero are considered as under-expressed miRNAs (green), while log2FC greater than zero represents over-expressed miRNAs (red). The miRNA patterns of each cancer type vary from each other. B. Counts of cancer types in which each miRNA is over- or under-expressed - vertical axis stands miRNAs (same as Figure 2A). Green bars represent the counts of cancer types in which a miRNA is under-expressed, and red ones are for cancer type counts in which a miRNA is over-expressed. After unsupervised clustering, some miRNAs show the same trend of over-expression or underexpression across most cancers. It suggests that these miRNAs may have similar function across different cancer types. C. The vertical axis represents the average of log2FC values generated from DESeq2 and edgeR for each miRNA in each cancer type. For each cancer, all of its miRNAs that are significantly differential expressed are included. The numbers of over- and under-expressed miRNAs per cancer vary from each other, suggesting the up-regulation of oncogenes is more important in the occurrence of some cancers, while the down-regulation of tumor suppression genes is more important in other cancers.