Figure 7: mTORC1 activation and substrate phosphorylation are highest at the lysosomal surface.
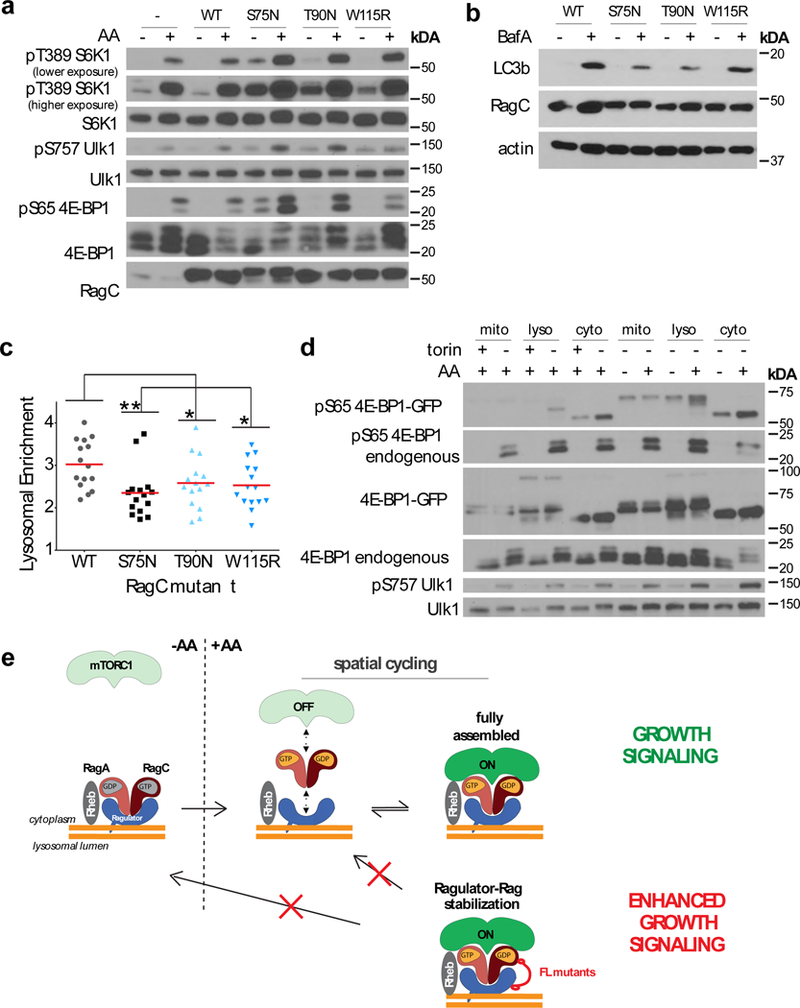
(a)HEK-293T cells stably expressing Flag-RagC harboring the indicated mutations were starved for amino acids for 50 min or starved and restimulated for 10 min, lysed, and immunoblotted for the indicated proteins and phosphor-proteins. Experiment repeated 3 times. Unprocessed scans are shown in Supplementary Fig. 8. (b) HEK-293T cells stably expressing Flag-RagC containing the indicated mutations were treated with vehicle or BafA for 2h, then lysed and immunoblotted for the indicated proteins. Experiment performed 1 time. Unprocessed scans are shown in Supplementary Fig. 8. (c) HEK-293T cells stably expressing Flag-RagC containing the indicated mutations were treated with BafA for 2h, fixed and immunostained for LC3 and LAMP2, then quantitated for the Lysosomal Enrichment Score of LC3 signal at LAMP2-positive lysosomes versus cytoplasmic LC3 signal (mean ± S.D., N = 15 cells/condition, **p = .0036, (T90N) *p = 0.0498, (W115R) *p = 0.0230, two-sided unpaired t-tests). Experiment performed 1 time. (d) HEK-293T cells stably expressing GFP-4E-BP1-OMP25 (mito), GFP-4E-BP1-Rheb15 (lyso) or GFP-4E-BP1-Rheb15-CAAX mutant (cyto) were treated with torin overnight or with torin overnight followed by a 2-hour torin washout (lanes 1-6), or were starved of amino acids for two hours or starved then restimulated for 10 minutes (lanes 7-12), followed by cell lysis and immunoblotting for the indicated proteins and phosphor-proteins. Experiment repeated 2 times. Unprocessed scans are shown in Supplementary Fig. 8. (e) Model for nutrient-induced mTORC1 capture to the lysosome. In low-nutrient states, Rag heterodimers loaded with the inactive nucleotide combinations are stably bound to Ragulator on the lysosomal surface. Nutrients cause the switch of the Rags to the active nucleotide state, while destabilizing Rag interaction with Ragulator. Consequently, both Rag GTPases and mTORC1 undergo spatial cycling, which limits the pool of mTORC1 activated by Rheb at the lysosomal surface. Cancer-specific RagC mutants decrease the off-rate of the Rag-Ragulator interaction and stabilize mTORC1 at the lysosome, resulting in increased signaling. Stabilization of the Rag-Ragulator interaction may also prevent Rag inactivation as nutrient levels decrease, maintaining mTORC1 in a constitutively active state. See Supplementary Table 1 for statistical source data.
