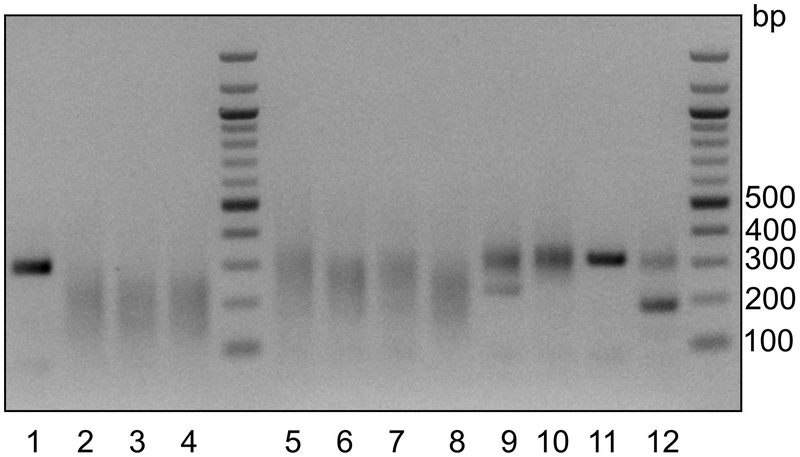
Figure 5. scGESTALT barcode editing.
scGESTALT barcode zebrafish were crossed to zebrafish that express heat shock-inducible Cas9 and U6-driven sgRNAs 5–9. Resulting embryos were injected with Cas9 protein and sgRNAs 1–4 at the one-cell stage. Embryos were heat shocked at 30 hpf to induce transgenic Cas9 for a late round of editing. Double transgenic (scGESTALT+, hsp:Cas9+; lanes 2–8, n = 7 embryos) and single transgenic (scGESTALT+, hsp:Cas9−; lanes 9–12, n = 4 embryos) were identified by screening for GFP expression. The gel shows PCR results of amplifying the scGESTALT barcode (unedited = ~300 bp). Large smear patterns (120–250bp) are observed in early and late edited embryos (lanes 2–8), whereas embryos that were only mutated at sites 1–4 display less editing (lanes 9–12. The band at ~200 bp in lane 12 likely represents large deletion(s) between sites 1–4 that occurred early in development and was inherited by most cells. Note that samples with such dominant large deletions should not be used for downstream experiments and analyses as they are likely to have low barcode diversity). Sample in lane 11 was likely not efficiently injected. Lane 1 represents a control embryo, which was injected with Cas9 protein only (no sgRNAs 1–4, n = 1 embryo) and was not heat shocked. As expected, the barcode is not edited in this case. This procedure was approved by the HU/FAS Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08.