Figure 1.
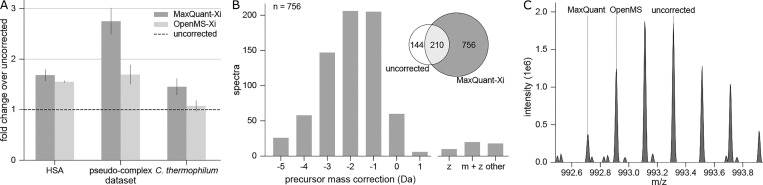
Correction of the monoisotopic peak is crucial in cross-link identification. (A) The data sets were preprocessed using MaxQuant and OpenMS, leading to more identified PSMs in all cases. Fold changes from uncorrected data (msconvert conversion of Xcalibur data) were calculated for each file separately and the mean plotted. Error bars represent the standard error of the mean between different acquisitions (HSA: n = 3, pseudocomplex: n = 3, C. thermophilum: n = 8). (B) The majority of additional identifications after preprocessing are due to correction of the precursor mass to lighter monoisotopic masses. Spectra that are unique to MaxQuant preprocessed searches of HCD acquisitions from data set 2 were evaluated in terms of precursor correction. The main proportion of the gain was corrected to lighter masses of up to −3 Da, while charge state correction or correction to heavier masses rarely occurred. (C) Isotope cluster of a corrected precursor of m/z 992.71 (z = 5, m = 4958.6 Da) was solely identified in MaxQuant preprocessed results. In OpenMS preprocessed and uncorrected data, the wrong monoisotopic mass was selected for unknown reasons.