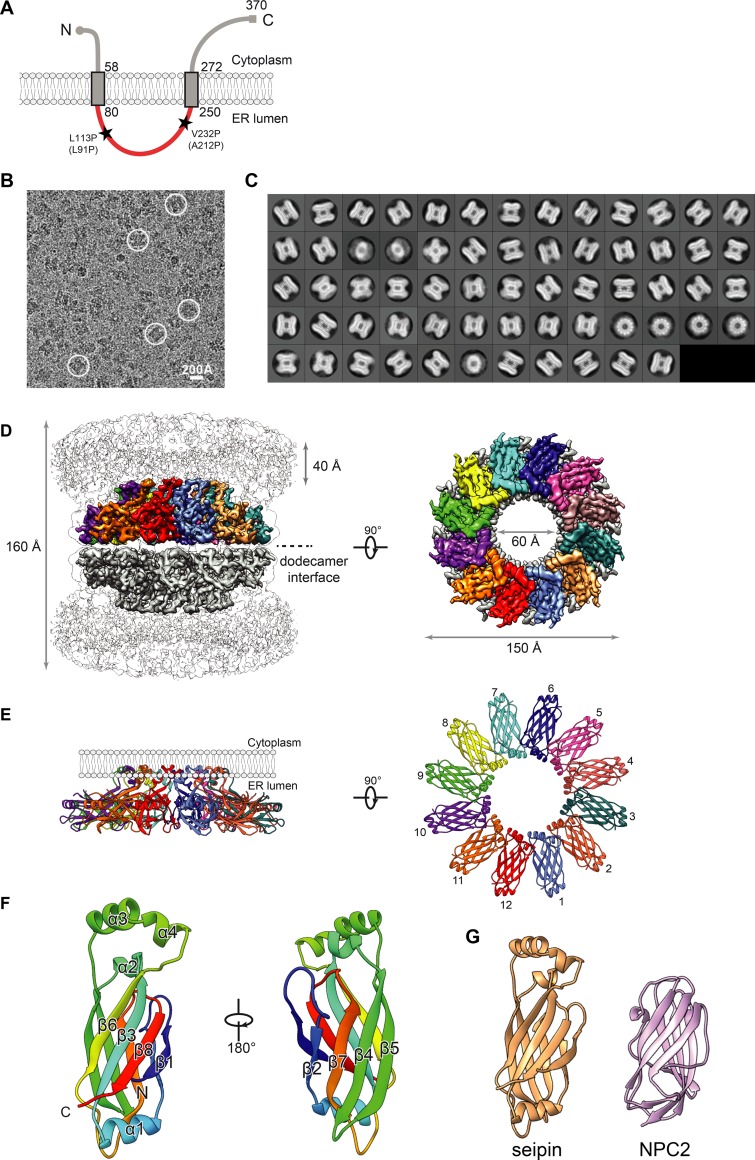
Figure 1.
Cryo-EM map and molecular model of seipin. (A) Model of D. melanogaster seipin topology in the ER membrane. The conserved ER-luminal domain is in red. Human pathogenic mutations L91P and A212P and their equivalent positions in D. melanogaster seipin are shown. (B) Representative cryo-EM image of purified seipin in digitonin. White circles indicate representative particle images. (C) 2D averages calculated with all seipin particles from combined datasets. The box dimension is 335 Å. (D) Unsharpened (transparent) and sharpened (solid colored) cryo-EM density maps of a seipin oligomer. The barrel-like structure is a head-to-head dimer of dodecamers interacting via luminal domains. The 40-Å region at the top represents poorly resolved TM regions. Each monomer in the upper dodecamer ring is shown in different colors. The en face view is shown from the perspective of the ER membrane. (E) Ribbon diagram side (left) and top (right) views of the luminal domains. (F) Model and structural elements of seipin monomers containing residues 88–240 corresponding with the ER luminal domain. Each domain contains a β sandwich of eight β strands and four α helices. Helices 3 and 4 comprise hydrophobic sequences positioned at the ER luminal leaflet. (G) Structural comparison of seipin (orange) and the cholesterol-binding protein NPC2 (pink; PDB accession ID: 2HKA); mean root mean square displacement of 4.3 Å over 106 residues.