Figure 3.
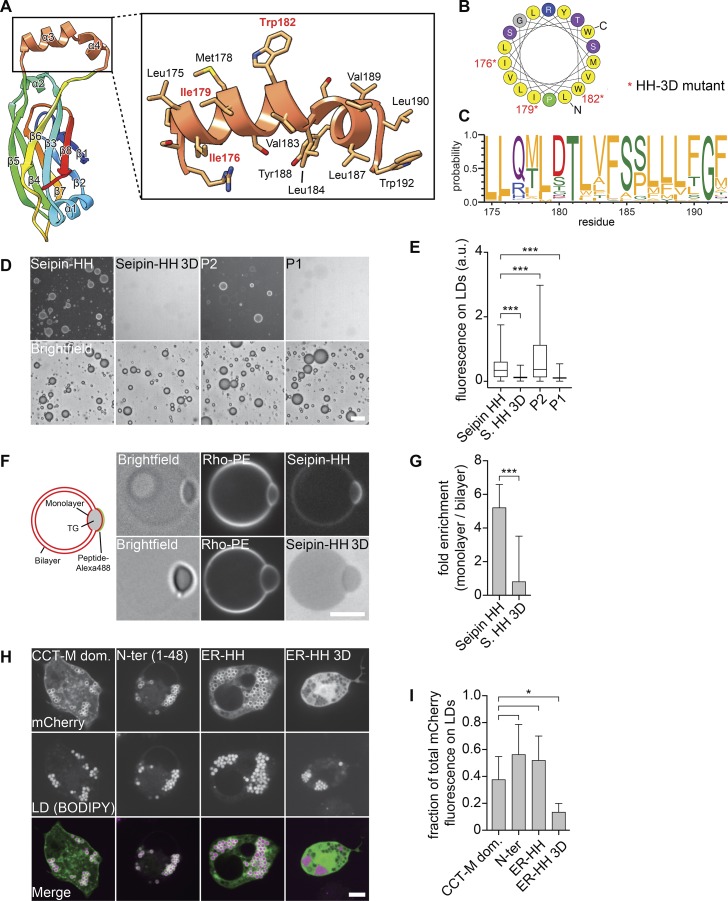
The HH of the seipin ER luminal domain targets to LDs. (A) Molecular structure of the HH highlighting residues 172–192 in orange. (B) Helical plot of residues Leu175–Trp192. Nonpolar residues are shown in yellow (Gautier et al., 2008). Asterisks indicate residues mutated to Asp in the seipin 3D mutant. (C) The helical region residue distribution for the top 200 seipin sequences (retrieved from the Pfam database; corresponding with residues 175–192 of D. melanogaster seipin) shows evolutionary conservation of hydrophobicity (Crooks et al., 2004). Residues are colored according to their physicochemical properties, with hydrophobic residues in orange. (D) The seipin HH binds artificial LDs in vitro. An Alexa Fluor 488–labeled peptide comprising residues 174–193 but not a version with the 3D mutation (replacing Ile176, Ile179, and Trp182 with Asp) binds to artificial LDs. (E) Quantification of fluorescent signals from >2,000 artificial LDs per peptide as in C as a boxplot representation. (F) The seipin HH binds to the phospholipid monolayer in vitro. Seipin helix peptide but not the mutated 3D version preferentially binds to the monolayer of TG lenses incorporated into GUVs. Graphical representation and representative confocal images show the peptide and phospholipid signals, respectively. (G) Quantification of enrichment on a monolayer versus a bilayer of ≥18 GUVs per peptide as mean ± SD. (H) Binding of seipin HH to LDs in cells. The mCherry-tagged N-terminal amphipathic helical sequence (1–48), the luminal HH (174–193) of seipin, and a seipin-HH 3D mutant of the luminal helix were expressed in D. melanogaster S2 cells and analyzed by confocal imaging for LD binding. As a control, the CTP-phosphocholine cytidylyltransferase (CCT) M domain was expressed in S2 cells from the same vector. Bars: 20 µm (D); 5 µm (F and H). (I) Quantification of mCherry fluorescence on LDs versus total signal per cell as shown in H as mean ± SD from ≥9 cells per construct. *, P < 0.01; ***, P < 0.0001.