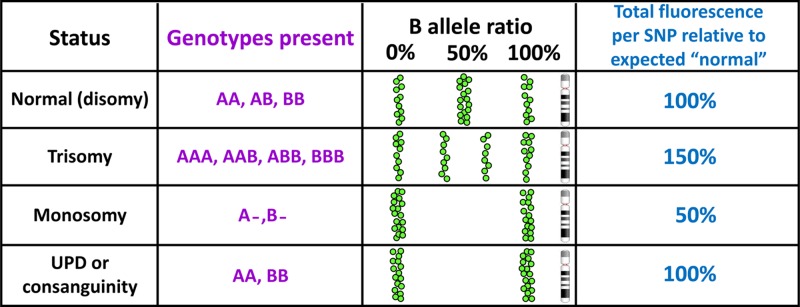
Figure 46. SNP array data can reveal copy number imbalance and homozygosity associated with consanguinity and UPD.
For ease of display and interpretation in SNP arrays the two alleles of each SNP are conventionally designated A and B, thus, for a T/C SNP, T would become the ‘A’ allele and C would become the ‘B’ allele. Each SNP is genotyped and the result plotted (as a green spot) according to position along the chromosome in terms of ‘B allele ratio’, which will be 0% for AA, 50% for AB and 100% for BB. Trisomy is revealed by B allele frequencies of 33 and 66%, and by an overall gain in fluorescence. In monosomy there is only one allele for each SNP so there will be no heterozygosity, and the total fluorescence will be only half the expected value. Long runs of homozygosity resulting from UPD or consanguinity will generate normal fluorescence levels. Other states can also be diagnosed, for example mosaicism (mixtures of two or more cell lines) will lead to skewed B allele ratios.