Figure 6. Comparison of motifs generated by different approaches for three HLA-DR alleles.
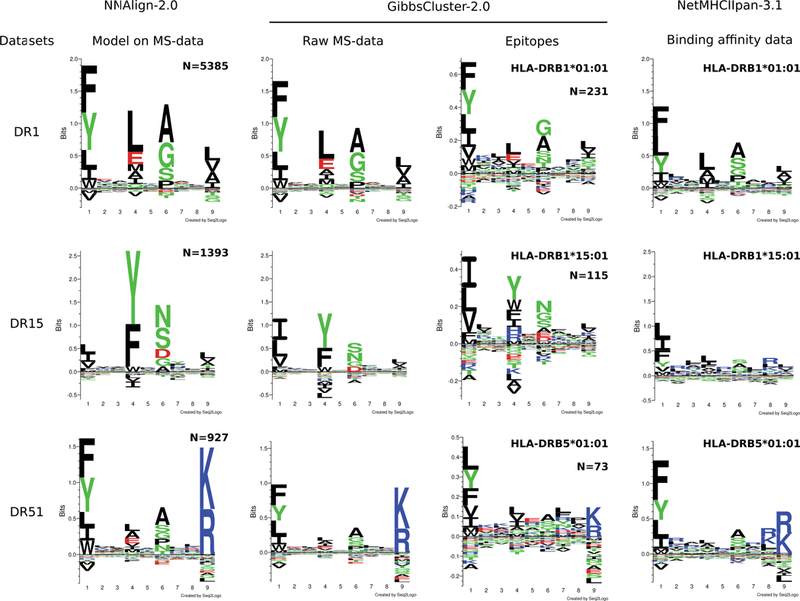
NNAlign-2.0 motifs were obtained by training artificial neural networks on each MS data set, and evaluating 100,000 random peptides. The top scoring 1% peptides were used to build logos. Raw MS data were aligned, clustered and filtered in an unsupervised manner using GibbsCluster, with a trash cluster threshold = 2. The same procedure was applied to tetramer-positive data downloaded from the IEDB. Note that due to small data set size, epitope logos are shown in a different y-axis scale. Binding motifs for NetMHCIIpan-3.1 were determined by evaluating 100,000 random peptides, and visualizing the core motif of the top 1% scoring sequences.
