Figure 3.
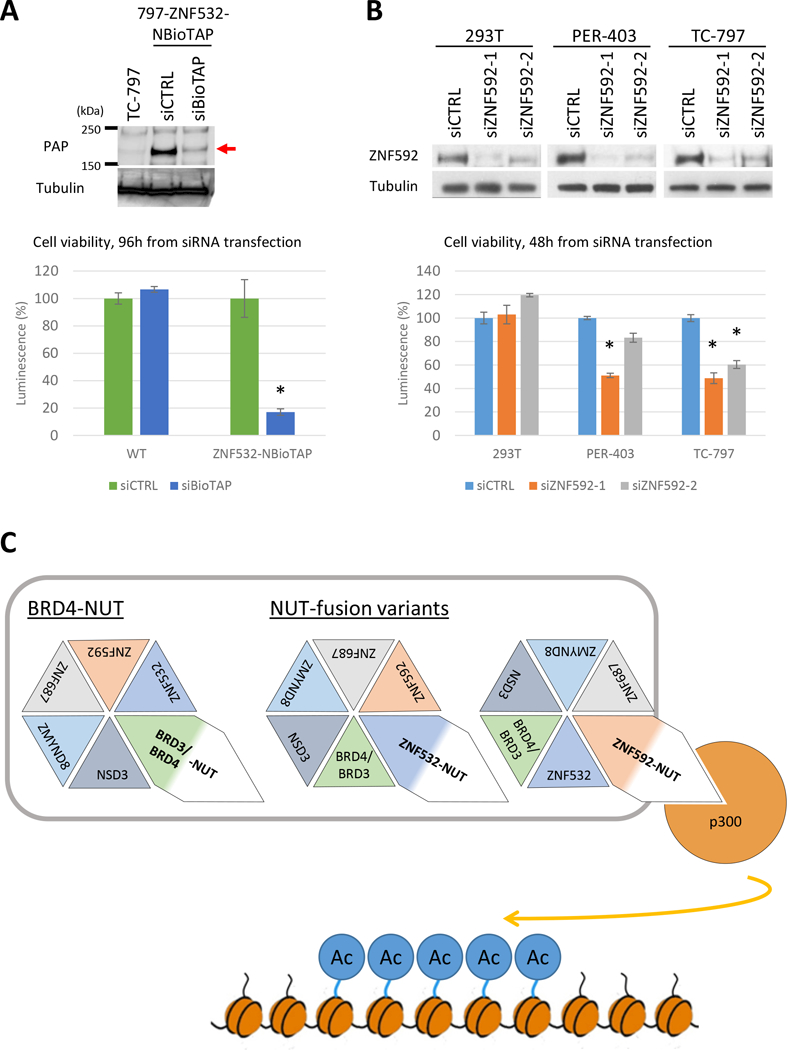
ZNF592 and ZNF532 are critical for NC cell viability. A, Top: Immunoblot of TC-797, 797-ZNF532-NBioTAP at 48 hours after siCTRL or siBioTAP using Peroxidase Anti-Peroxidase (PAP) antibody, which binds the protein A moiety of BioTAP and visualizes ZNF532-NBioTAP protein on the blot. ZNF532-NBioTAP protein band is indicated by a red arrow. Tubulin was used as a loading control. Bottom: Viability assays performed on TC-797 or 797-ZNF532-NBioTAP cells at 96 hours following transfection with siCTRL or siBioTAP. 8000 cells were plated per well in a 96-well plate. Error bars represent standard deviation (SD) from three biological replicates. *p<0.01, student’s t test. B, Top: Immunoblot of 293T, PER-403 and TC-797 at 48 hours after transfected with siCTRL, siZNF592–1 or siZNF592–2 with ZNF592 antibody. Tubulin was used as a loading control. Bottom: Viability assay performed on TC-797, PER-403 and 293T cells 48 hours following transfection with siCTRL, siZNF592–1 or siZNF592–2. 8000 cells were plated per well in a 96-well plate. Error bars represent SD from three biological replicates. *p<0.01, student’s t test. C. Mechanistic model of how NUT-fusions function. Associations of NUT, BET protein and p300 through NUT-fusions are essential to drive proliferation and blockade of differentiation by inducing ectopic histone hyperacetylation on broad region of the chromatin (“megadomain”). This association is mediated by interaction of NUT-fusion protein with other associated proteins (BRDs, ZNF532, ZNF592, ZMYND8, ZNF687, NSD3) or formation of BRD3/4-NUT, NSD3-NUT (not shown here), ZNF532-NUT or ZNF592-NUT fusion.
