Abstract
Background & Aims
Hepatitis C virus (HCV) co-opts the very low-density lipoprotein (VLDL) pathway for morphogenesis, maturation, and secretion, and circulates as lipoviroparticles (LVPs). We investigated the functions and underlying mechanisms of the lipid-associated transmembrane 6 superfamily 2 (TM6SF2) protein in modulating LVP formation and the HCV life cycle.
Methods
We knocked down or overexpressed TM6SF2 in hepatic cells and examined HCV infection, measuring viral RNA and protein and infectious LVP. The density of secreted LVPs was evaluated by iodixanol gradient assay. We measured levels and patterns of TM6SF2 in liver biopsies from 73 patients with chronic hepatitis C, livers of HCV infected humanized Alb-uPA/SCID mice, and HCV-infected Huh7.5.1 cells.
Results
TM6SF2 knockdown in hepatocytes reduced viral RNA and infectious viral particle secretion without affecting HCV genome replication, translation, or assembly. Overexpression of TM6SF2 reduced intracellular levels of HCV RNA and infectious LVPs, and conversely increased their levels in the culture supernatants. In HCV-infected cells, TM6SF2 overexpression resulted in production of more infectious LVPs in the lower-density fractions of supernatant. HCV infection increased TM6SF2 expression in cultured cells, humanized livers of mice, and liver tissues of patients. TM6SF2 mRNA levels correlated positively with HCV RNA levels in liver biopsies from patients. SREBP-2 appears to mediate the ability of HCV to increase the expression of TM6SF2 in hepatic cells.
Conclusion
In studies of cells, mice and human liver tissues, we found TM6SF2 is required for maturation, lipidation and secretion of infectious LVPs. HCV, in turn, upregulates expression of TM6SF2 to facilitate productive infection.
Keywords: Infectious Hepatitis C virus particles, release, lipid metabolism
Introduction
Hepatitis C virus (HCV) circulates in the form of lipoviroparticles (LVPs), which contain assembled viral particles and very low-density lipoprotein (VLDL) components consisting of apolipoproteins B, E, and C1 (ApoB, ApoE, and ApoC1), triglycerides and cholesteryl ester.1–5 LVPs that have low densities (<1.06 g/mL) are highly infectious, implicating that lipidation is important in maturation and release of infectious virions. Several previous studies have proposed that HCV exploits the VLDL synthetic pathway for assembly, maturation and secretion.6–10 Nevertheless, the precise mechanism of HCV LVP morphogenesis remains elusive and needs to be identified.11 HCV glycoproteins are associated with ApoB and ApoE on the endoplasmic reticulum (ER) membrane.10 While some studies demonstrated that LVPs require functional microsomal triglyceride transfer protein (MTP) and ApoB to gain infectivity,8,9 others suggested that these factors are dispensable and only ApoE is important in the generation of infectious viral particles.12–15 Indeed, after nucleocapsid (NC) formation near the ER, HCV neo-particles will transit along the VLDL maturation pathway where they acquire lipid components, thus generating infectious LVPs with heterogeneous buoyant densities.16,17
To elucidate global HCV–host interactions, our laboratory recently conducted functional genomics screens and identified a large number of previously unappreciated host factors that modulate various stages of the HCV life cycle.18,19 Among them, transmembrane 6 superfamily member 2 (TM6SF2) represents a pivotal host dependency for HCV infection.19 Interestingly, TM6SF2 is also associated with other hepatic disorders. In several genome-wide association studies, its E167K variant was shown to accelerate the intracellular degradation of TM6SF2 and confer a 2.1-fold higher risk of non-alcoholic fatty liver disease (NAFLD).20 TM6SF2 E167K carriers also demonstrated lower circulating levels of total cholesterol, low-density lipoprotein (LDL), and triglyceride (TG) than non-carriers.21 In several TM6SF2-depleted models of mice or cultured cells, the secretion of TG and cholesterol was decreased, leading to hepatic steatosis manifested by an increase of lipid droplet (LD) contents in hepatocytes.22–25 In addition, upon TM6SF2 knockdown, a modest decrease of ApoB export could be observed in multiple hepatocyte cell lines,25 but this effect was not seen in TM6SF2 KO mice.24
Intriguingly, chronic hepatitis C (CHC) patients carrying the E167K variant of TM6SF2 are associated with a higher risk of hepatic steatosis and fibrosis, and lipid abnormalities.26–30 Thus, it is important to define the precise functions of TM6SF2 in modulating HCV infection and associated metabolic liver diseases.
In this study, we identified TM6SF2 as a critical host factor mediating HCV secretion/release. We demonstrated that it enhances neo-LVPs maturation, lipidation and secretion. We also showed that TM6SF2 expression is significantly upregulated and positively correlates with HCV RNA levels in liver biopsies of CHC patients, HCV-infected SCID/uPA mouse livers and HCV-infected cultured hepatocytes. Finally, we showed that SREBP-2, a key transcriptional regulator of cholesterol metabolism31,32 that is induced in HCV-infected hepatocytes,33,34 is responsible for the enhanced transcription of TM6SF2 mRNA upon HCV infection.
Material and Methods
Patient and mouse samples
Patient samples were provided by the Liver Clinic at the National Institutes of Health Clinical Center from a large cohort of chronic hepatitis C (CHC) patients. Available liver biopsies from 73 HCV infected patients were used for analysis of hepatic TM6SF2 expression. Because of sample limitation, a subgroup of the samples (n = 29) with TM6SF2 rs58542926 genotype C/C (E167) were used to compare the TM6SF2 mRNA levels between HCV-infected and normal liver tissues, and to evaluate correlation between TM6SF2 mRNA and hepatic HCV RNA levels. All patients provided written informed consent for participation in clinical research and genetic testing. Normal human liver tissues (frozen, n=19) were provided by the National Institutes of Health–supported Liver tissue Cell Distribution system. Generation of the Albu-PA/SCID/beige mice and transplantation of primary human hepatocytes (PHHs) derived from the same donor were performed as described.35 Mice were injected intravenously with 50 μL of HCV-positive (genotype 1b, 105 genomes) patient serum as described.35 For human gene expression measurements, the specificity of human primer probes has been tested with wild type mice materials and has no cross-reactivity with murine mRNA.36
Proteolytic digestion protection assay
As previously described,37 and detailed in the Supplementary Material and Methods section.
Kinetic of HCV RNA and LVP secretion
The siRNA transfected cells were infected with HCV for 24 h, and then incubated with medium supplemented with sofosbuvir (10 μM) and anti-CD81 Ab (10 ng/mL) for 2, 4, 6, 8 and 10 h. HCV RNA were determined for each time point from 0 h to 10 h. For intracellular infectivity assays, infected cells were washed with PBS, and lysed by incubation in 200 μL of water to induce hypo-osmotic shock. Lysates were clarified by centrifugation at 10,000 x g for 10 minutes at 4°C. 100 μL of each cell lysate was used to infect Huh7.5.1 cells. Infection was examined at 48 h p.i. by qRT-PCR with HCV primers/probes and normalized to 18S RNA. The relative expression was normalized to the value of siNT sample.
Density gradient
Discontinuous 5–50% iodixanol density gradients (OptiPrep, Sigma-Aldrich, St. Louis, MO) were prepared from five buffered solutions of iodixanol in HEPES-buffered saline (Sigma-Aldrich, St. Louis, MO) as previously described.38 Samples were overlaid onto the gradients and centrifuged for 24 h at 4°C and 50,000 rpm in a SW60Ti rotor in a Beckman Coulter Optima L-90 K ultracentrifuge (Brea, CA). Fractions (250 μl each) were collected and their amount of HCV RNA and infectivity and their density were determined respectively by viral RNA isolation and quantification and by measuring absorbance at 340 nm. We determined the infectivity of the viral preparation, by infecting naïve Huh7.5.1 cells (100,000 per well in a 12-well plate) with 25 μL of each virus fraction and incubating for 48 h at 37°C. Total RNA was extracted. HCV-RNA levels were measured by qRT-PCR in using a standardized HCV RNA quantification panel from AcroMetrix. We estimated the infectivity by calculating the relative HCV infectious unit (RIU) that is defined as the ratio between intracellular HCV RNA level at 48 h p.i. and the inoculum HCV RNA level in 25 μL of the fraction.
TM6SF2 genotyping
446 patients with chronic hepatitis C who were seen at the NIH Clinical Center Liver Clinic and provided consent for genetic testing (protocol 91-DK-0214) were genotyped for the rs58542926 SNP, a nonsynonymous variant in exon 6 of TM6SF2. Genotyping was determined by TaqMan assay (Genome Quebec, McGill University, Quebec, Canada) and was successful in 445 patients (99%). Serum HCV RNA levels prior to any treatment were evaluated for association with the TM6SF2 genotype by Linear regression analysis with JMP genomics 7.0 (SAS Institute Inc, Cary, NC).
Results
TM6SF2 is requisite for infectious HCV lipoviroparticle production.
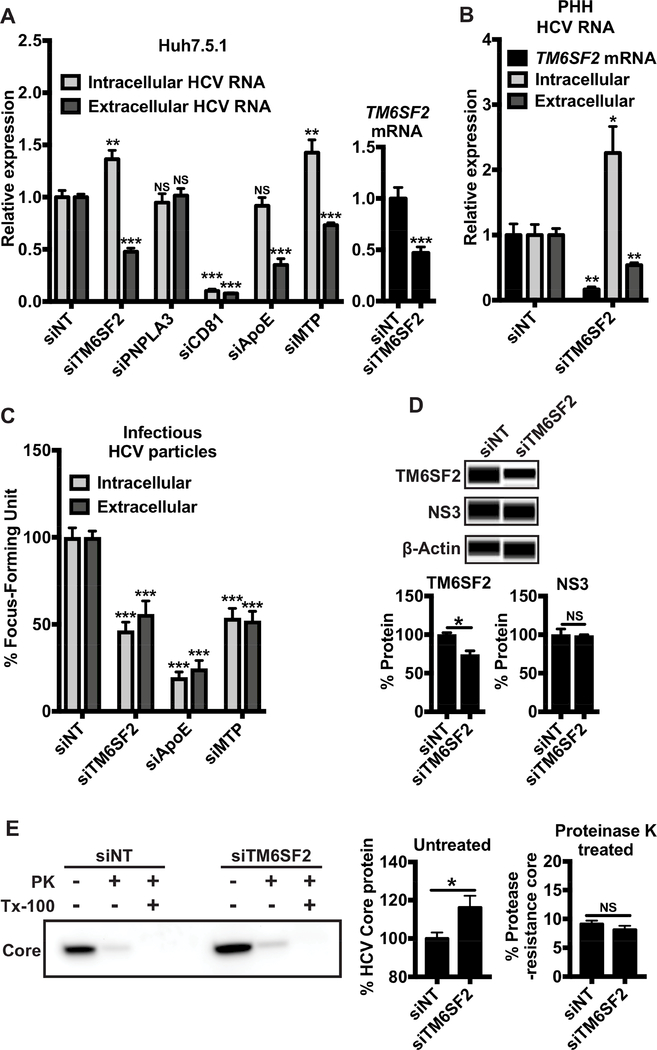
In a previous genome-wide RNAi screen, we showed that TM6SF2 is involved in the HCV life cycle but does not affect either viral entry or replication.19 To define the precise roles of TM6SF2 in modulating HCV infection, we knocked down its expression in Huh7.5.1 cells and primary human hepatocytes (PHHs). In Huh7.5.1 cells, TM6SF2 mRNA level was decreased to about 50% by siTM6SF2 without obvious cytotoxicity (Fig. 1A). We evaluated the impacts of TM6SF2 knockdown on HCV RNA and infectious lipoviroparticles (LVPs) production. After siRNA-mediated TM6SF2 depletion and at 48 h p.i., we extracted total RNA from cell lysates or viral supernatants. HCV RNA levels were quantified by quantitative real-time reverse transcriptase-PCR (qRT-PCR) and normalized to non-targeting control siRNA (siNT). siRNAs targeting PNPLA3 (siPNPLA3) or CD81 (siCD81) were used as a negative or positive control, respectively (Fig. S1). In Huh7.5.1 cells and PHHs, TM6SF2 silencing resulted in 1.5- and 2-fold increase of intracellular HCV RNA levels, respectively, and decreased extracellular HCV RNA levels for 2-fold in both cell culture models (Fig. 1A, B). Intracellular viral RNA levels also increased in siTM6SF2-transfected cells infected with chimerical HCV of various genotypes (Fig. S2). The downregulated TM6SF2 functions can be rescued by the transfection of a TM6SF2 ectopic expression vector but not by an ApoB overexpression vector (Fig. S3A, B). The effects of TM6SF2 silencing on HCV RNA levels were similar to those of MTP knockdown but somewhat different from those of ApoE knockdown (no increase in intracellular HCV RNA levels)(Fig. 1A, S1). ApoE and MTP are involved in the assembly and maturation of infectious LVPs with distinct mechanisms.16
Figure 1: TM6SF2 knockdown decreases the production and secretion of HCV infectious particles.
Huh7.5.1 or PHHs cells were transfected with siRNA as indicated for 72 h and then infected with HCV. Total RNAs were extracted from cells at 48 h p.i. In (A) Huh7.5.1 (n=9) or (B) PHHs (n=5), the relative expressions of HCV RNA or TM6SF2 mRNA were measured in total RNA extractions after quantification of HCV RNA by qRT-PCR (standardized to 18S RNA). (C) Intra- and extracellular infectious particles were evaluated by incubate the cell lysate and supernatant respectively, of cells with naïve Huh7.5.1 cells for 48 h. Cells were stained with anti-HCV core Ab and viral foci were counted manually. The numbers of viral foci were normalized to that of siNT control and shown as % Focus Forming Units, 3 independent experiments in triplicates (n=9). (D) Total proteins were extracted at 48 h p.i. Proteins were detected using Western blot (Wes™ system), with the indicated antibodies. The peak of chemiluminescence was obtained by Compass software and the % protein expression was normalized to that of the siNT control (100%) (n=4). (E) Proteolytic digestion protection assays of lysates were performed on HCV infected cells depleted of TM6SF2 or siNT control, demonstrating the amount of protease-resistant core following no treatment or treatment with Triton (Tx-100) and Proteinase K. Plotted data represent relative core levels from 3 independent experiments in triplicates (n=9) quantified by Western blotting. In the proteinase K untreated samples, the TM6SF2 knockdown values were normalized to those of the siNT control (middle panel). In the proteinase K treated samples, the core levels of the non-targeting and TM6SF2 knockdown samples were normalized to those of untreated siNT and siTM6SF2 samples, respectively (100%) (right panel). Shown values are means ± SEM. NS, non-significant; *, P-value < .05; **, P-value < .01; ***, P-value < .001 (Mann Whitney’s test).
We then measured HCV infectious titers (FFUs) from the supernatants and cell lysates of above-examined cells. Both the intracellular and extracellular levels of infectious HCV decreased for about 50% upon TM6SF2 knockdown, an effect comparable to MTP knockdown (Fig. 1C). TM6SF2 is not involved in HCV RNA replication (Fig. S4,19). The expression level of the nonstructural protein 3 (NS3), which reflects viral replication and protein synthesis, was not altered by TM6SF2 silencing (Fig. 1D); yet the level of HCV core protein was increased (Fig. 1E, left and middle panels), suggesting that TM6SF2 may act at the late stages of HCV infection. We then evaluated the level of fully enveloped capsid by examining the resistance of core to proteinase K (PK) digestion as previously described.37 Lysates from HCV-infected, TM6SF2 knockdown or NT control cells were treated with or without 1% Triton (Tx-100), followed by PK digestion. Residual core protein was quantified by Western blotting. Treatment with PK alone resulted in core proteolysis in both samples, with a minimal fraction of core remaining protected from the protease (Fig. 1E). Thus HCV envelopment during viral assembly does not seem to be affected by TM6SF2 knockdown. These results suggest that TM6SF2 may act at post-assembly steps of the HCV life cycle, such as maturation and/or secretion.
Next we knocked down TM6SF2 in Huh7.5.1 cells and analyzed the levels of triglycerides and cholesteryl ester – two major components of LDs, and the size of the LDs (Fig. S5). In these cells, the LD-positive area, and triglycerides and cholesteryl ester contents were significantly increased (Fig. S5). These findings are consistent with previous reports.20,25
We tried to generate a complete TM6SF2 KO Huh7.5.1 cell line by CRISPR/Cas9 editing. We were only able to generate a partial knockdown clone (one-allele), suggesting that cells with a complete knockout of TM6SF2 may not be viable.
TM6SF2 promotes HCV RNA and infectious lipoviroparticle secretion.
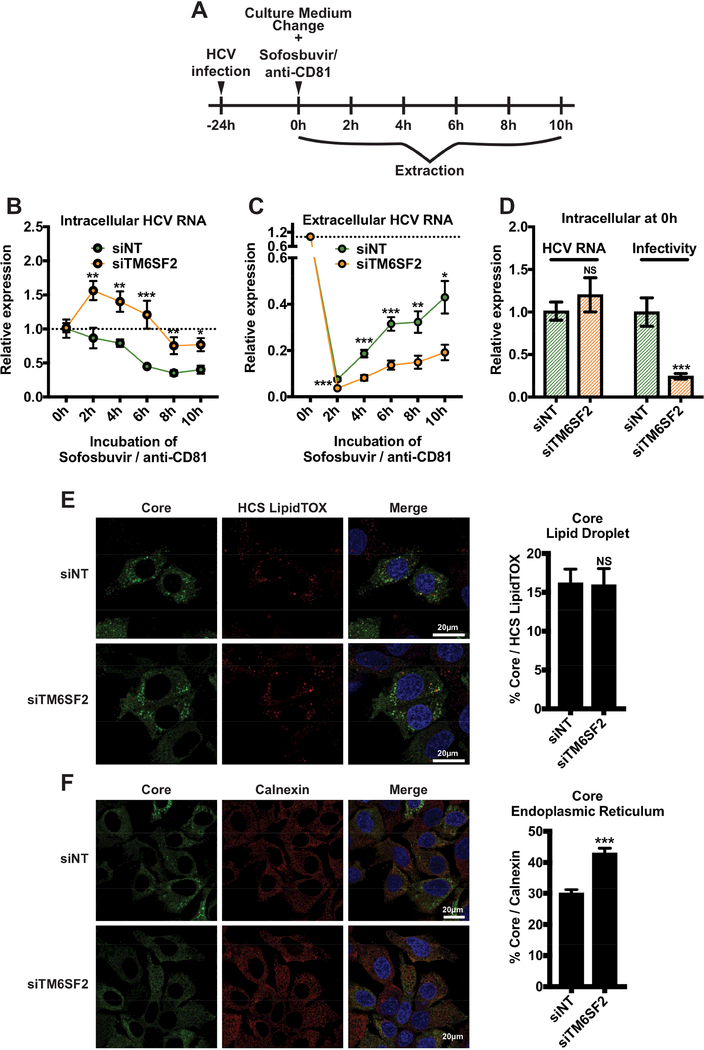
Through loss-of-function assays, we demonstrated that TM6SF2 is required for production of infectious LVPs but has no effect on viral replication, protein translation or assembly. To determine whether TM6SF2 is involved in HCV maturation and/or secretion, we examined the kinetics of HCV RNA secretion upon TM6SF2 depletion. siNT or siTM6SF2-treated cells were infected with HCV for 24 h and then treated with sofosbuvir to terminate viral replication39 and together with anti-CD81 antibody to block new infection.40 Samples were collected at various time points after drug treatment (Fig. 2A). Intra and extracellular HCV RNA levels were quantified by qRT-PCR and normalized to the level at 0 h under each treatment condition. With this method, we can trace HCV RNA that was already synthetized. To evaluate intracellular infectivity, we collected cell lysate at 0 h post-treatment and used them to infect naïve cells. At 48 h p.i., total cellular RNA was extracted, and HCV RNA levels were determined by qRT-PCR and normalized to 18S RNA. Relative HCV infectivity was calculated by comparing to control cells (siNT). We showed that for siNT, the levels of intracellular HCV RNA progressively decreased with time, reaching a 2-fold reduction at 6 h post-treatment comparing to that of 0 h (Fig. 2B). In contrast, the extracellular HCV RNA levels were increasing with time (Fig. 2C). These data indicate that while viral replication was blocked by drug treatment, the assembly and secretion of viral RNA were unaffected. In TM6SF2-silenced cells, the intracellular HCV RNAs were generally higher at various time points than those of siNT-treated cells (Fig. 2B). Interestingly, the HCV RNA first showed a 1.5-fold increase after the addition of sofosbuvir and anti-CD81, and afterwards started to decline (Fig. 2B). The transient increase of intracellular HCV RNA level can be explained by the continuous effect of TM6SF2 knockdown on the HCV life cycle before sofosbuvir and anti-CD81 were fully active in blocking viral infection. On the other hand, the levels of extracellular HCV RNA upon TM6SF2 knockdown were significantly lower than those secreted from control cells at various time points (Fig. 2C). These data implicate that TM6SF2 deletion interferes with HCV secretion.
Figure 2: TM6SF2 knockdown blocks the secretion of HCV RNA and infectious particles.
Huh7.5.1 cells were transfected with siRNA as indicated for 72 h and infected with HCV. (A) 24 h post-infection, at time point 0 h, media were changed and supplemented with sofosbuvir and anti-CD81 Ab. Samples were harvested at 2, 4, 6, 8 and 10 h after treatment. (B) Intracellular and (C) extracellular total RNA were extracted and HCV RNA were quantified of by qRTPCR (standardized to 18S RNA). (D) At time point 0 h, the relative expression of HCV RNA was quantified and cell lysates (50 μL) were used to infect naïve Huh7.5.1 cells. At 48 h p.i., total RNAs were then extracted and HCV RNA levels were quantified by qRT-PCR to establish HCV infectivity. HCV RNA and infectivity were compared to the siNT condition. All experiments were independent and performed 3 times in triplicates (n=9). (C) 48 h p.i. cells are fixed, HCV core, LDs (HCS LipidTOX) and ER (anti-calnexin Ab) were stained. Percentages of colocalization between core and LDs or ER were quantified, using Zen software (at least n=64). White bars scale, 20 μm. Shown values are means ± SEM. NS, non-significant; *, P-value < .05; **, P-value < .01; ***, P-value < .001 (Mann Whitney’s test).
At 24 h p.i., before the treatment with sofosbuvir and anti-CD81 antibody, the levels of HCV RNA were comparable between siNT and siTM6SF2-transfected cells, whereas the intracellular infectivity was considerably lower in TM6SF2-silenced cells (Fig. 2D). These results confirmed the inhibitory effect of TM6SF2 knockdown on the production of infectious LVPs as observed above (Fig. 1A, C).
HCV assembly starts with the association of core around the LDs followed by the budding of the neo-assembled viral particles inside the ER.16 With our kinetic experiment we established that HCV secretion is dependent on TM6SF2. To determine in which compartment the viral particles are retained, we doublelabeled HCV core protein and LDs or ER (calnexin as a marker) in infected, siNT or siTM6SF2-transfected cells and evaluated the percentage of colocalization under each condition. In siTM6SF2-treated cells, HCV core colocalization with ER was significantly enhanced; but not with the LDs (Fig. 2E, F). These results suggest TM6SF2 regulates HCV secretion after core assembly in the ER compartment.
TM6SF2 facilitates lipidation of lipoviroparticles.
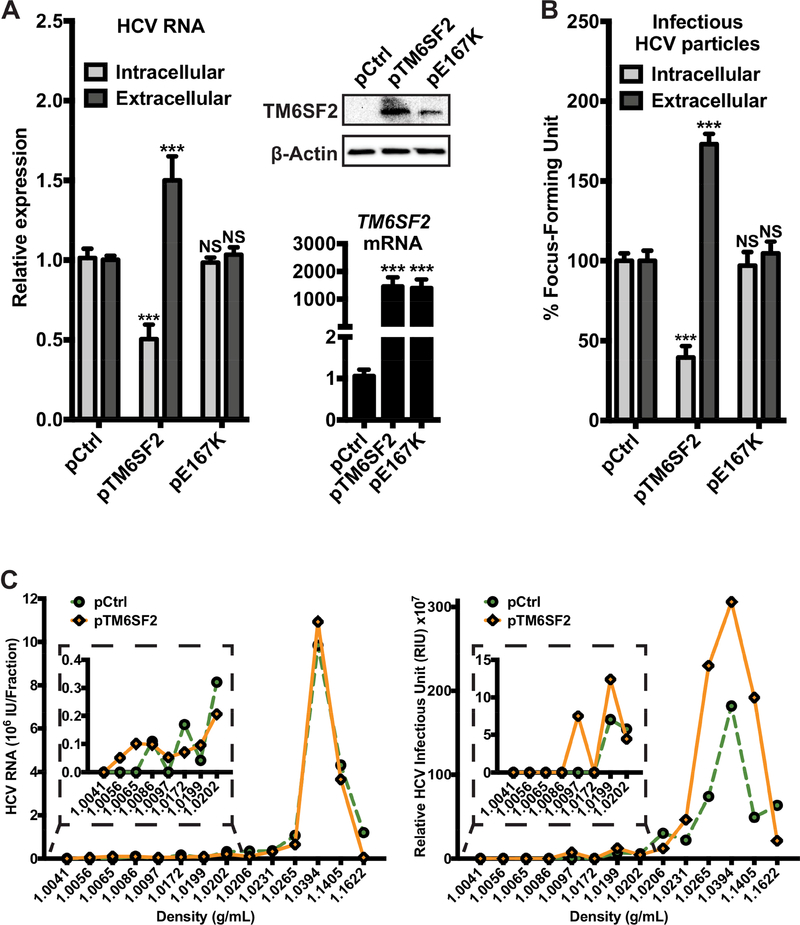
To further explore the role of TM6SF2 in modulating HCV infection, we conducted gain-of-action assays in Huh7.5.1 cells overexpressing a plasmid carrying the TM6SF2 gene (pTM6SF2). Eight hours after plasmid transfection, cells were infected with HCV. At 48 h p.i., we measured the expression levels of HCV RNA and the FFU levels. The levels of HCV RNA and infectious LVPs showed similar alteration pattern when TM6SF2 is overexpressed (Fig. 3A, B). There was an around 2-fold decrease in intracellular HCV RNA and infectious LVPs levels, whereas both levels in the supernatant were increased for about 1.5-fold (Fig. 3A, B). The decrease in intracellular HCV RNA and infectious LVPs levels comparing with an increase in extracellular levels suggest that more assembled viral RNA and LVPs were secreted to the medium.
Figure 3: Overexpression of functional TM6SF2 increases the secretion of HCV RNA and infectious lipoviroparticles.
Huh7.5.1 cells were transfected with TM6SF2 (pTM6SF2) or the mutant TM6SF2-E167K (pE167K) expression plasmids for 8 h and infected with HCV. Total RNA and proteins extractions were obtained from those transfected cells at 48 h p.i. (A) Relative expressions of HCV RNA and of TM6SF2 were measured in total RNA extractions after detection of HCV RNA or TM6SF2 mRNA by qRT-PCR (standardized to 18S RNA). Proteins were detected by Western blot with the indicated antibodies. (B) The intra- and extracellular infectious particles were evaluated by inoculating the cell lysate and supernatant respectively, to naïve Huh7.5.1 cells for 48 h. Cells were then stained with anti-HCV core Ab and viral foci were counted manually. The numbers of viral foci were normalized to that of siNT control and shown as % Focus Forming Units. A and B are a plot of 3 independent experiments in triplicates (n=9). Shown values are means ± SEM. ***, P-value < .001 (Mann Whitney’s test). (C) The supernatants from HCV-infected Huh7.5.1 cells overexpressing TM6SF2 or control cells were subjected to isopycnic centrifugation through iodixanol gradients, as described in Materials and Methods. For each fraction, total RNA was extracted and HCV RNA were quantified of by qRT-PCR and presented by on a graph (left panel). 25 μL of each fraction was used to infect Huh7.5.1 cells. At 48 h p.i., total RNA was then extracted and HCV RNA levels were quantified by qRT-PCR. The infectivity, expressed in relative HCV infectious unit (RIU), was defined as the ratio between the intracellular HCV RNA level 48 h p.i. and the inoculum HCV RNA in 25 μL of each fraction (right panel).
The secretion of infectious HCV particles relies on the VLDL pathway,16 and TM6SF2 has been shown to facilitate lipid loading onto lipoproteins.23,24 The secreted LVPs have heterogeneous densities due to there varying extents in complex with luminal lipids during HCV maturation and secretion. It has been suggested that LVP density is determined by its lipid content,41 and that lower LVP density (higher lipidation) is associated with increased infectivity.42 To characterize the role of TM6SF2 in the lipidation of LVPs, we overexpressed the protein and analyzed the density of secreted HCV RNA and LVPs in the supernatant.
Huh7.5.1 cells were transfected with TM6SF2 or control plasmid before infection with HCV. The viral supernatants were subsequently subjected to centrifugation through a gradient of buoyant densities. The HCV RNA and infectivity of LVPs was evaluated per each fraction (Fig. 3C). LVPs from both supernatants showed HCV RNA and infectivity at buoyant densities of IDL and LDL (1.019 g/mL to 1.063 g/mL and 1.006 g/mL to 1.019 g/mL, respectively43), which are consistent with previous reports.3,44 Fractions 1.0394 g/mL to 1.1622 g/mL represent those of the samples loaded on the gradient. Interestingly, supernatants of pTM6SF2-transfected cells contained infectious LVPs from fraction 1.0097 to 1.0265 g/mL, while infectious LVPs in the control supernatant were present from fraction 1.0199 to 1.0265 g/mL (Fig. 3C). In low buoyant density fraction (such as 1.0097 g/mL), infectious LVPs were detected only in the supernatant of pTM6SF2-transfected cells (Fig. 3C, insert). On the other hand, secreted LVPs from the supernatant of TM6SF2-depleted cells were absent at the buoyant densities of IDL and LDL (Fig. S6). Thus, we conclude that TM6SF2 is involved in the lipidation of infectious HCV particles.
TM6SF2-E167K variant and HCV infection.
The TM6SF2-E167K variant has been shown to impair the expression and function of the gene.22 We thus examined the effect of this variant on HCV infection. Overexpression of the TM6SF2-E167K variant resulted in the same level of TM6SF2 mRNA as the wildtype but the protein level was lower. The decreased protein level in association with the E167K variant has been reported previously, possibly attributed to a lower stability of the protein.20 In contrast to the wild-type, overexpression of the E167K variant did not affect HCV RNA levels or production and secretion of LVPs (Fig. 3A, B, S3A). In a cohort of CHC patients (Table S1), we did not observe any significant association of the TM6SF2 rs58542926 SNP (C>T, E167K variant) with the serum HCV RNA levels (Fig. S3C).
Induction of TM6SF2 mRNA and protein expression during HCV infection.
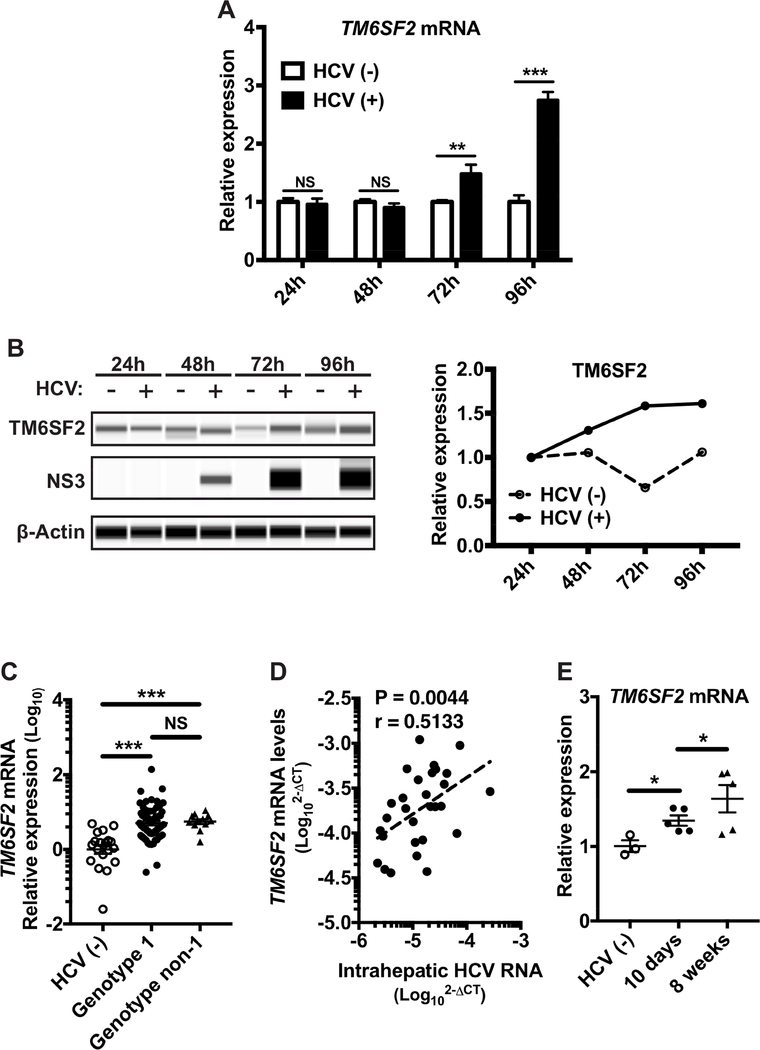
To further elucidate TM6SF2–HCV interactions, we examined the level of TM6SF2 transcripts during HCV infection. In HCV-infected Huh7.5.1 cells, TM6SF2 mRNA and protein levels were significantly elevated from 72 h p.i. (Fig. 4A, B). In liver biopsies of CHC patients with TM6SF2 rs58542926 genotype C/C (E167), TM6SF2 mRNA levels were significantly higher than those in liver tissues of healthy controls, independent of HCV genotypes (Fig. 4C). Interestingly, TM6SF2 mRNA levels positively correlated with HCV RNA levels in these liver biopsies (Fig. 4D). On the other hand, hepatic TM6SF2 mRNA levels did not correlate with HCV viremic levels (Fig. S7). HCV viremic levels depend on many factors other than HCV production by the infected hepatocytes. Thus it is not surprising that hepatic HCV RNA levels correlate better with TM6SF2 expression. Moreover, in Alb-uPA/SCID/beige mice transplanted with human hepatocytes, HCV infection led to a gradual increase of hepatic TM6SF2 levels, coinciding with progressive HCV spread (viremia typically peaks at 3–6 weeks post-infection45) (Fig. 4E).
Figure 4: Increased expression of TM6SF2 in cell line, liver biopsies of HCV infected patients and mouse models.
(A) Total RNA extractions were obtained from HCV infected Huh7.5.1 cells at 24, 48, 72 and 96 h p.i. (B) Total proteins were extracted from infected Huh7.5.1 cells at 24, 48, 72 and 96 h p.i. Proteins were detected by Western blot (Wes™ system), with the indicated antibodies. The peak of chemiluminescence was obtained by Compass software and normalized to the level at 24 h for each time point. (C) Total RNA extractions were obtained from liver biopsies of HCV infected patients (HCV(−): n=19; HCV genotype 1: n=61; genotype non-1: n=12) and the relative expression of TM6SF2 mRNA was measured after detection of TM6SF2 mRNA by qRT-PCR (standardized to 18S RNA). (D) The correlation (Spearman rank correlation’s test) between HCV RNA and TM6SF2 mRNA (2-∆CT) was measured after quantification by qRT-PCR (standardized to 18S RNA) in a sub-group of 29 liver biopsies. All HCV patients analyzed here carry the CC (major allele) genotype. (E) Total RNA extractions were obtained from HCV infected humanized AlbuPA/SCID/beige mouse’s livers. The relative expression of TM6SF2 mRNA was shown relative to that of TBP after quantification of TM6SF2 and TBP mRNAs by qRT-PCR (standardized to 18S RNA). Shown values are means ± SEM. NS, non-significant; *, P-value < .05; **, P-value < .01; ***, P-value < .001 (Mann Whitney’s test).
SREBP-2 mediates TM6SF2 induction during HCV infection.
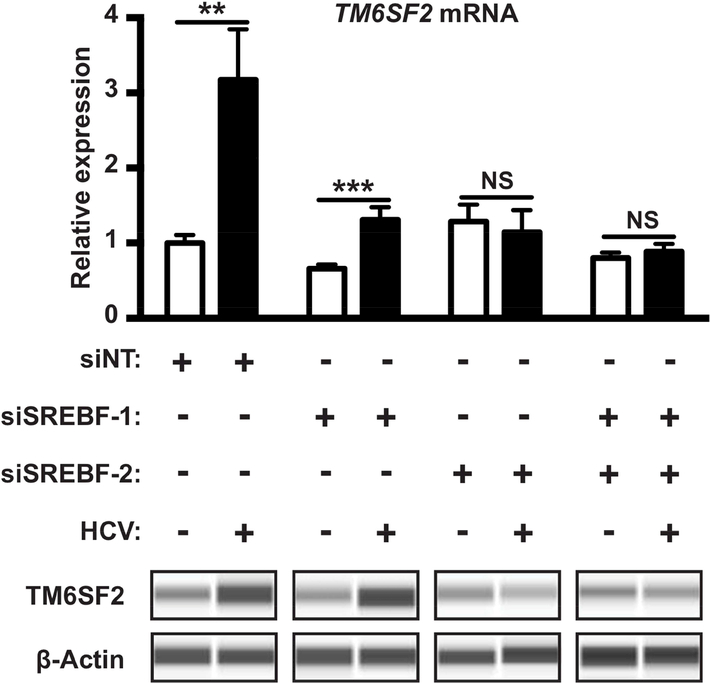
TM6SF2 regulates lipid metabolism and HCV infection but how its own expression is regulated is not entirely clear.23–25 The sterol regulatory element-binding proteins (SREBPs) including SREBP-1 and SREBP-2 are master transcriptional regulators of cholesterol and fatty acid metabolism, respectively.31,32 SREBPs have been shown to be induced and activated in HCV-infected hepatocytes.33,34 To investigate whether the increase of TM6SF2 expression during HCV infection is SREBP-dependent, we knocked down SREBP-1 or SREBP-2 or both by siRNAs (Fig. S1) and examined TM6SF2 expression either in the absence or in the presence of HCV infection. When SREBP-1 was depleted, a significant increase of TM6SF2 mRNA and protein levels by HCV infection was still observed as compared to non-infected control. However, in SREBP-2 depleted cells, the levels of TM6SF2 mRNA and protein were comparable between the infected and the uninfected cells (Fig. 5). Similar phenotypes were observed in SREBP-1/2 dual knockdown cells (Fig. 5). These data suggest that SREBP-2 but not SREBP-1 is involved in TM6SF2 induction during HCV infection.
Figure 5: SREBP-2 mediates increased TM6SF2 expression during HCV infection.
Huh7.5.1 cells were transfected with SREBF-1 siRNA, SREBF-2 siRNA or both for 72 h, and then infected with HCV. Total RNAs and proteins were obtained from those transfected cells at 96 h p.i. The relative expressions of TM6SF2 mRNA were determined in total RNAs after quantification of by qRTPCR and normalization with 18S RNA. Plotted data represent relative expression from 3 independent experiments in triplicates (n=9). Shown values are means ± SEM. NS, non-significant; **, P-value < .01; ***, P-value < .001 (Mann Whitney’s test). Proteins were detected by Western blot (Wes™ system), with the indicated antibodies.
Discussion
We have previously shown that TM6SF2 is a proviral host factor during HCV infection.19 Our current study revealed that TM6SF2 manipulation in hepatocytes affects the maturation and secretion of HCV viral particles. In TM6SF2-silenced cells, there was a significant increase of intracellular HCV RNA, albeit TM6SF2 had no effect on HCV replication or translation (Fig. 1, S4,19). We then showed that the secretion of HCV RNA and infectious particles was reduced when TM6SF2 is partially depleted, subsequent to a diminished production of mature LVPs. Thus, we speculate that loss of TM6SF2 hampers the maturation and secretion of HCV particles, leading to an accumulation of newly synthesized and assembled HCV RNA in the ER of the cells. Under this condition, though viral RNAs are properly packaged for assembly, the LVP maturation process is deficient. On the other hand, when TM6SF2 is overexpressed, levels of HCV RNA and infectious viral particles decreased in the cells but increased in the medium. We reason that TM6SF2 overexpression facilitates neo-LVPs maturation and secretion at the post-assembly stage, thereby elevating the extracellular HCV particle levels. Enhanced secretion of LVPs thus results in diminished intracellular levels of HCV RNA and infectious viral particles.
Our data showed similar effect of TM6SF2 or MTP knockdown on intra and extracellular HCV RNA levels (Fig 1A). The function of MTP in HCV life cycle is controversial8,9,12–15 but our results suggest that MTP, like TM6SF2, may play an important role in HCV maturation and secretion.
HCV hijacks the process of lipoprotein production for LVP assembly, thereby the lipid composition of LVPs is important for the maintenance of viral infectivity. Depleting their cholesterol or TG contents increases viral particle density and thus leading to loss of HCV infectivity.42 In our study, when TM6SF2 was overexpressed, more infectious LVPs appeared to be in lower buoyant densities than the negative control group, supporting that TM6SF2 is involved in the lipidation, maturation and/or secretion of LVPs.
Several previous studies suggested that TM6SF2 is important in the lipidation and/or export of VLDL.22,24,25 Moreover, a genetic polymorphism of TM6SF2 (E167K) confers a 2.1-fold higher risk of non-alcoholic fatty liver disease (NAFLD) and this variant accelerates TM6SF2 intracellular degradation, thus decreasing its expression.20 In both mouse models and cultured cells, TM6SF2 downregulation leads to hepatic steatosis manifested by increased TG contents in hepatocytes, decreased total cholesterol and TG lipoprotein export, and reduced size of VLDL particles due to a defect in the lipidation process.22–25 In our present study, as expected, overexpression of the TM6SF2-E167K variant did not alter the secretion of HCV RNA or LVPs. We conclude that HCV requires a fully functional TM6SF2 protein for maturation and/or secretion. In contrast, TM6SF2 overexpression increased circulating total cholesterol and TG contents in the mouse model47 and decreased LD contents in Huh7 cells.25 TM6SF2 is also thought to be a sterol isomerase in the cholesterol metabolism pathway.48 Thus, there is an intimate interaction between TM6SF2 and lipid metabolism. The complete function of TM6SF2, nevertheless, remain to be fully elucidated. In CHC patients, association of the TM6SF2-E167K variant with HCV viremic levels has been controversial.26–30 Our genotyping data in a cohort of 393 CHC patients did not show any significant association.
We showed that HCV infection increases TM6SF2 expression in cultured hepatocytes and Alb-uPA/SCID/beige mouse livers. TM6SF2 mRNA levels also correlate positively with intrahepatic HCV RNA levels in livers of HCV patients. We and others have shown previously that during HCV infection, key lipogenic enzymes and transcription factors SREBP-1/2 are induced and activated.33,46 Here we demonstrated that SREBP-2, a master regulator of cholesterol metabolism in cells,44,45 is primarily responsible for the increased expression of TM6SF2, suggesting that TM6SF2 is part of the cholesterol metabolic machinery.48 It is possible that pathways other than that mediated by SREBP-2 may also be involved in the regulation of TM6SF2 expression. The upregulation of TM6SF2 may represent a previously unidentified strategy for HCV to harness the cellular environment and host dependencies for its propagation.
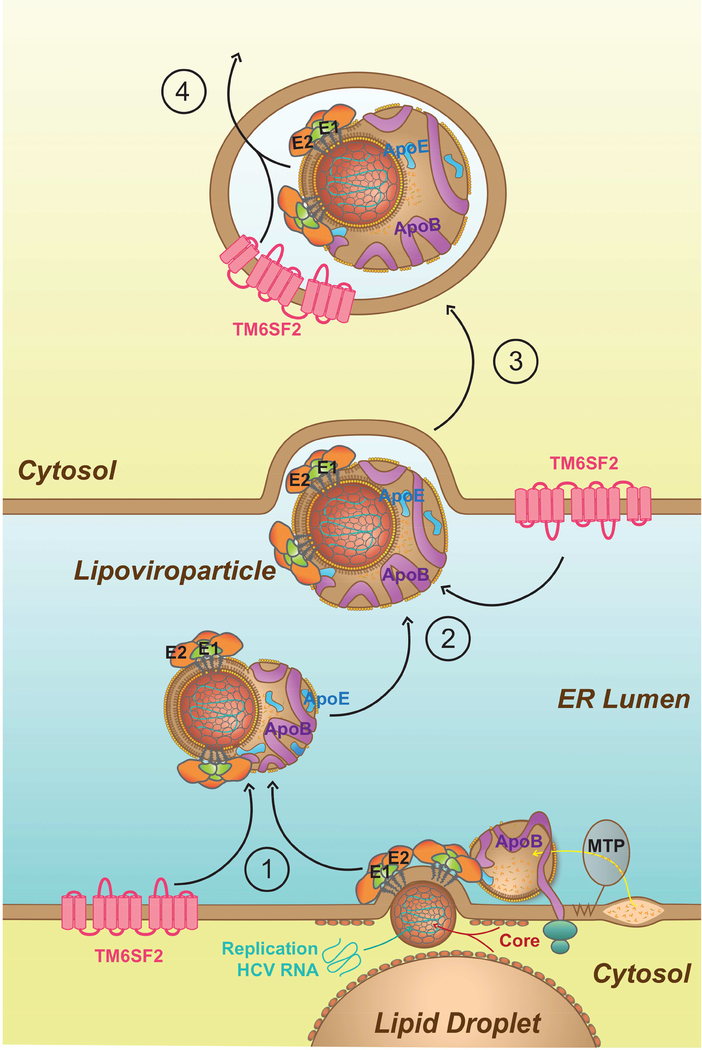
In summary, we demonstrated that TM6SF2 is a pivotal host dependency factor for HCV infection. TM6SF2 facilitates the lipidation of newly assembled LVPs and thus enhances their secretion (see model in Fig. 6). On the ER membrane, HCV glycoproteins complex with apolipoproteins B and E that trigger viral budding.10 TM6SF2 is localized to the ER and Golgi compartments,24 and first loads lipids unto nascent LVPs during the fusion between HCV particle and lipoprotein in the ER lumen (Fig. 6, step 1). Next, in the ER lumen, TM6SF2 may continue to enhance the LVPs lipidation to increase their infectivity (step 2). Lipidation by TM6SF2 may occur further during viral secretion (step 3), leading to a heterogenic population of secreted HCV particles wherein the most infectious LVPs have the lowest densities.42 Finally, TM6SF2-mediated lipidation is closely linked to secretion of infectious virions (step 4), which is similar to that of lipoprotein maturation and secretion.16 HCV, on the other hand, induces the expression of TM6SF2 to its advantage by enhancing the production of infectious virus.
Figure 6: Proposed Model for the Function of TM6SF2 in HCV LVP lipidation and secretion.
During HCV budding inside the ER lumen, apolipoproteins B and E associate with the enveloped virus. Once released into the ER, the nascent virion acquires all the components of lipoprotein (apolipoproteins and lipids) to produce LVP, the infectious form of HCV. Together with MTP, TM6SF2 may facilitate lipid loading onto the newly assembled virion during the budding and/or maturation of LVP (1). After the formation of infectious LVP, TM6SF2 may also enrich the lipid composition of HCV particles in the ER lumen (2) and/or during the secretory pathway through the Golgi apparatus or secretory vesicles (3). The extent of lipidation by TM6SF2 may be linked to viral infectivity and an important signal for efficient secretion of LVP (4).
Supplementary Material
Acknowledgment
The authors would like to specially thank Dr. Maren Podszun for her support during the project. We thank Kazuyaki Chayama and Xiaoming Cheng for providing mouse liver samples and reagents; Yanling Ma, Pierre-Christian Violet and Antony Cougnoux for their advice. Normal human liver tissues were obtained through the Liver Tissue Cell Distribution System (Minneapolis, MN), which was funded by a National Institutes of Health contract (HHSN276201200017C).
Financial Support: This research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Abbreviations
- Apo
Apolipoprotein
- CHC
chronic hepatitis C
- HCV
hepatitis C virus
- ER
endoplasmic reticulum
- FFU
foci forming unit
- LD
lipid droplet
- LDL
low density lipoprotein
- LVP
lipoviroparticles
- MTP
microsomal triglyceride transfer protein
- NAFLD
non-alcoholic fatty liver disease
- NC
nucleocapsid
- NT
Non-Targeting
- PHH
Primary Human Hepatocytes
- siRNA
small interfering RNA
- SREBP
sterol regulatory element-binding protein
- TG
triglyceride; p.i, post infection
- TM6SF2
transmembrane 6 superfamily member 2
- VLDL
very low density lipoprotein
- qRT-PCR
quantitative real-time reverse transcriptase-PCR
Footnotes
Conflicts of interest: The authors disclose no conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Thomssen R, Bonk S, Propfe C, et al. Association of hepatitis C virus in human sera with beta-lipoprotein. Med Microbiol Immunol (Berl) 1992;181:293–300. [DOI] [PubMed] [Google Scholar]
- 2.Prince AM, Huima-Byron T, Parker TS, et al. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J Viral Hepat 1996;3:11–17. [DOI] [PubMed] [Google Scholar]
- 3.André P, Komurian-Pradel F, Deforges S, et al. Characterization of Low- and Very-Low-Density Hepatitis C Virus RNA-Containing Particles. J Virol 2002;76:6919–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merz A, Long G, Hiet M- S, et al. Biochemical and Morphological Properties of Hepatitis C Virus Particles and Determination of Their Lipidome. J Biol Chem 2011;286:3018–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piver E, Boyer A, Gaillard J, et al. Ultrastructural organisation of HCV from the bloodstream of infected patients revealed by electron microscopy after specific immunocapture. Gut 2017;66:1487–1495. [DOI] [PubMed] [Google Scholar]
- 6.Wakita T, Pietschmann T, Kato T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 2005;11:791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gastaminza P, Kapadia SB, Chisari FV. Differential Biophysical Properties of Infectious Intracellular and Secreted Hepatitis C Virus Particles. J Virol 2006;80:11074–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gastaminza P, Cheng G, Wieland S, et al. Cellular Determinants of Hepatitis C Virus Assembly, Maturation, Degradation, and Secretion. J Virol 2008;82:2120–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H, Sun F, Owen DM, et al. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci 2007;104:5848–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer A, Dumans A, Beaumont E, et al. The Association of Hepatitis C Virus Glycoproteins with Apolipoproteins E and B Early in Assembly Is Conserved in Lipoviral Particles. J Biol Chem 2014;289:18904–18913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mankouri J, Walter C, Stewart H, et al. Release of Infectious Hepatitis C Virus from Huh7 Cells Occurs via a trans-Golgi Network-to-Endosome Pathway Independent of Very-Low-Density Lipoprotein Secretion. J Virol 2016;90:7159–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang J, Luo G. Apolipoprotein E but Not B Is Required for the Formation of Infectious Hepatitis C Virus Particles. J Virol 2009;83:12680–12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang K-S, Jiang J, Cai Z, et al. Human Apolipoprotein E Is Required for Infectivity and Production of Hepatitis C Virus in Cell Culture. J Virol 2007;81:13783–13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benga WJA, Krieger SE, Dimitrova M, et al. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology 2010;51:43–53. [DOI] [PubMed] [Google Scholar]
- 15.Da Costa D, Turek M, Felmlee DJ, et al. Reconstitution of the Entire Hepatitis C Virus Life Cycle in Nonhepatic Cells. J Virol 2012;86:11919–11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavie M, Dubuisson J. Interplay between hepatitis C virus and lipid metabolism during virus entry and assembly. Biochimie 2017;141:62–69. [DOI] [PubMed] [Google Scholar]
- 17.Popescu C-I, Riva L, Vlaicu O, et al. Hepatitis C Virus Life Cycle and Lipid Metabolism. Biology 2014;3:892–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Brass AL, Ng A, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci 2009;106:16410–16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Zhang Y-Y, Chiu S, et al. Integrative Functional Genomics of Hepatitis C Virus Infection Identifies Host Dependencies in Complete Viral Replication Cycle. PLOS Pathog 2014;10:e1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirola CJ, Sookoian S. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: A meta-analysis. Hepatology 2015;62:1742–1756. [DOI] [PubMed] [Google Scholar]
- 22.O’Hare EA, Yang R, Yerges-Armstrong LM, et al. TM6SF2 rs58542926 impacts lipid processing in liver and small intestine. Hepatology 2017;65:1526–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Y, Lu H, Guo Y, et al. Hepatic Transmembrane 6 Superfamily Member 2 Regulates Cholesterol Metabolism in Mice. Gastroenterology 2016;150:1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smagris E, Gilyard S, BasuRay S, et al. Inactivation of Tm6sf2, a Gene Defective in Fatty Liver Disease, Impairs Lipidation but Not Secretion of Very Low Density Lipoproteins. J Biol Chem 2016;291:10659–10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdessian H, Taxiarchis A, Popov S, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci 2014;111:8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coppola N, Rosa Z, Cirillo G, et al. TM6SF2 E167K variant is associated with severe steatosis in chronic hepatitis C, regardless of PNPLA3 polymorphism. Liver Int 2015;35:1959–1963. [DOI] [PubMed] [Google Scholar]
- 27.Milano M, Aghemo A, Mancina RM, et al. Transmembrane 6 superfamily member 2 gene E167K variant impacts on steatosis and liver damage in chronic hepatitis C patients. Hepatology 2015;62:111–117. [DOI] [PubMed] [Google Scholar]
- 28.Eslam M, Mangia A, Berg T, et al. Diverse impacts of the rs58542926 E167K variant in TM6SF2 on viral and metabolic liver disease phenotypes. Hepatology 2016;64:34–46. [DOI] [PubMed] [Google Scholar]
- 29.Sagnelli C, Merli M, Uberti-Foppa C, et al. TM6SF2 E167K variant predicts severe liver fibrosis for human immunodeficiency/hepatitis C virus co-infected patients, and severe steatosis only for a non-3 hepatitis C virus genotype. World J Gastroenterol 2016;22:8509–8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Que S, Zhou L, et al. The effect of the TM6SF2 E167K variant on liver steatosis and fibrosis in patients with chronic hepatitis C: a meta-analysis. Sci Rep 2017;7:9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madison BB. Srebp2: A master regulator of sterol and fatty acid synthesis1. J Lipid Res 2016;57:333–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X, So J-S, Park J-G, et al. Transcriptional Control of Hepatic Lipid Metabolism by SREBP and ChREBP. Semin Liver Dis 2013;33:301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su AI, Pezacki JP, Wodicka L, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci 2002;99:15669–15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Pène V, Krishnamurthy S, et al. Hepatitis C virus infection activates an innate pathway involving IKK-α in lipogenesis and viral assembly. Nat Med 2013;19:722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiraga N, Imamura M, Hatakeyama T, et al. Absence of viral interference and different susceptibility to interferon between hepatitis B virus and hepatitis C virus in human hepatocyte chimeric mice. J Hepatol 2009;51:1046–1054. [DOI] [PubMed] [Google Scholar]
- 36.Cheng X, Xia Y, Serti E, et al. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology 2017;66:1779–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentzsch J, Brohm C, Steinmann E, et al. Hepatitis C Virus p7 is Critical for Capsid Assembly and Envelopment. PLOS Pathog 2013;9:e1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maillard P, Walic M, Meuleman P, et al. Lipoprotein Lipase Inhibits Hepatitis C Virus (HCV) Infection by Blocking Virus Cell Entry. PLOS ONE 2011;6:e26637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatia HK, Singh H, Grewal N, et al. Sofosbuvir: A novel treatment option for chronic hepatitis C infection., Sofosbuvir: A novel treatment option for chronic hepatitis C infection. J Pharmacol Pharmacother J Pharmacol Pharmacother 2014;5, 5:278, 278–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fénéant L, Levy S, Cocquerel L. CD81 and Hepatitis C Virus (HCV) Infection. Viruses 2014;6:535–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartenschlager R, Penin F, Lohmann V, et al. Assembly of infectious hepatitis C virus particles. Trends Microbiol 2011;19:95–103. [DOI] [PubMed] [Google Scholar]
- 42.Fukuhara T, Ono C, Puig-Basagoiti F, et al. Roles of Lipoproteins and Apolipoproteins in Particle Formation of Hepatitis C Virus. Trends Microbiol 2015;23:618–629. [DOI] [PubMed] [Google Scholar]
- 43.Rader DJ, Hobbs HH. Chapter 356. Disorders of Lipoprotein Metabolism In: Longo DL, Fauci AS, Kasper DL, et al. , eds. Harrison’s Principles of Internal Medicine. 18th ed. New York, NY: The McGraw-Hill Companies; 2012. [Google Scholar]
- 44.Calattini S, Fusil F, Mancip J, et al. Functional and Biochemical Characterization of Hepatitis C Virus (HCV) Particles Produced in a Humanized Liver Mouse Model. J Biol Chem 2015;290:23173–23187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stacey AR, Norris PJ, Qin L, et al. Induction of a Striking Systemic Cytokine Cascade prior to Peak Viremia in Acute Human Immunodeficiency Virus Type 1 Infection, in Contrast to More Modest and Delayed Responses in Acute Hepatitis B and C Virus Infections. J Virol 2009;83:3719–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmen OL, Zhang H, Fan Y, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet 2014;46:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Pulido L, Ponting CP. TM6SF2 and MAC30, new enzyme homologs in sterol metabolism and common metabolic disease. Front Genet 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.