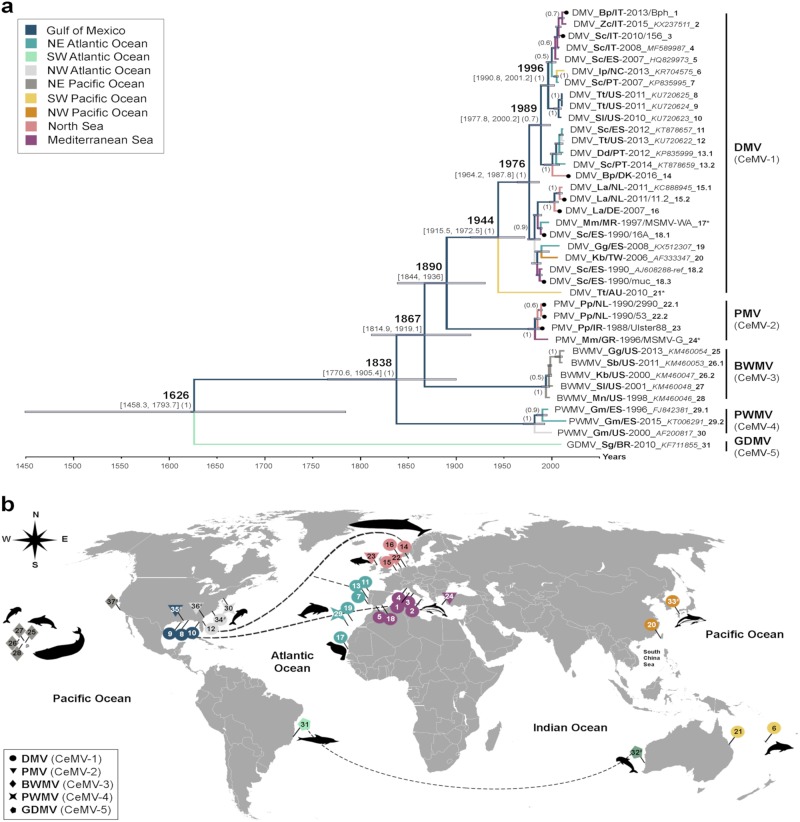
Fig. 3. Phylogeography of CeMV.
a Bayesian phylogenetic analysis of partial P genes (400 bp). Most common recent ancestor ages are presented at the nodes in cursive with 95% highest posterior density interval values in brackets and as gray horizontal bars. Posterior values > 0.5 are shown in parenthesis. CeMV genomes in this study are presented with black circles at the tips. Each branch is color-coded according to the ocean/sea in which the cetaceans were found. The taxon names are presented as virus_host/Country-year of collection/variant_GenBank accession number_ID for b. Sequences 17* and 24* were extracted from a published paper22. Sequence 21* was kindly provided by Dr. Stone and Jianning Wang. Host abbreviations: Bp Balaenoptera physalus, Dd Delphinus delphis, Gg Grampus griseus, Gm Globicephala melas, Ip Indopacetus pacificus, Kb Kogia breviceps, La Lagenorhynchus albirostris, Mm Monachus monachus, Mn Megaptera novaeangliae, Pp Phocoena phocoena, Sb Steno bredanensis, Sc Stenella coeruleoalba, Sg Sotalia guianensis, Sl Stenella longirostris, Tt Tursiops truncatus, and Zc Ziphius cavirostris. b A world map of CeMV migration using sequences from a. Locations of viruses in the map are relative and were obtained from the publications in which the sequences were published. The hashtag (#) indicates no sequence availability (No. 32: CeMV-5 from AU/2010 and No. 33: CeMV-1_JP/1999). Proposed virus migratory routes (dashed lines) were based on sequence clades from the phylogenetic tree in a