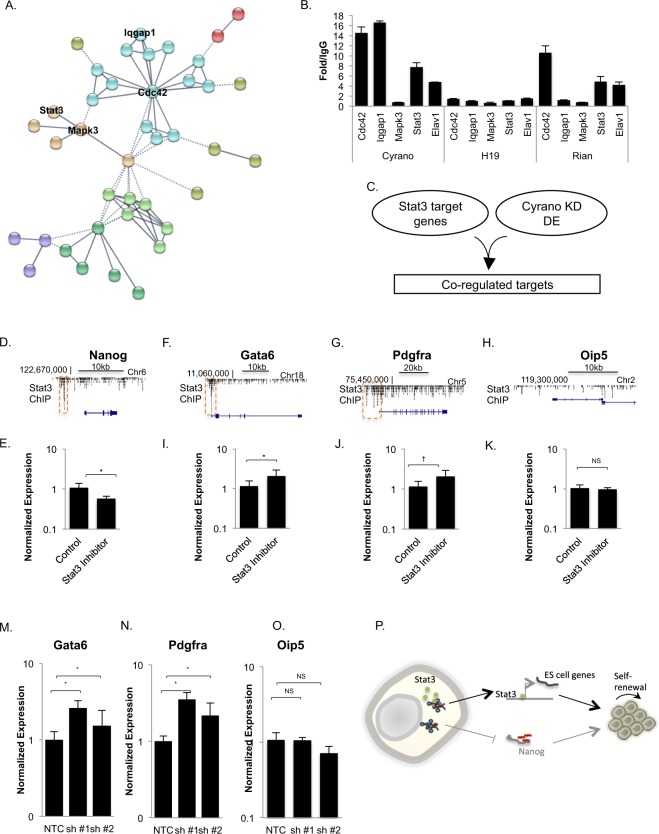
Figure 5.
Cyrano interacts in a Developmental/Signaling Hub. (A) A sub-network containing developmentally important proteins identified by CHART was selected for further study. RNA immunoprecipitation for select proteins, along with the RNA binding protein, ELAV1 (Table S2) was used as an independent assay to investigate Cyrano binding (B), relative to lncRNAs H19 and Rian. Correlation analysis between Stat3 target genes and genes differentially expressed (DE) upon Cyrano depletion was carried out (C). (D) UCSC genome browser (mm8) view of Stat3 ChIP-Seq peaks34 (dashed box) upstream of Nanog. (E) The Stat3 inhibitor Stattic was used to investigate the effect of inhibition on Nanog levels. (F,G) UCSC genome browser view (mm8) of Stat3 ChIP-Seq peaks34 upstream of Gata6 and Pdgfra respectively, relative to the non-target, Oip5 (H). (I–K) The Stat3 inhibitor Stattic was used to examine the effect of Stat3 inhibition on Gata6 and Pdgfra levels respectively, relative to Oip5. (M–O) The levels of Stat3 target, early lineage specification genes Gata6 and Pdgfra were examined upon Cyrano depletion using independent shRNAs (Dharmacon), relative to the Cyrano neighboring gene, Oip5. *p < 0.05, †p < 0.1. (P) Prospective model of an additional Cyrano function elucidated based on the mass-spectrometry determined interactome.