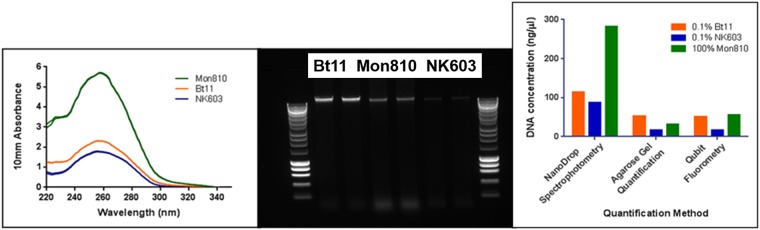
Figure 5.
Quantification of maize genomic DNA. Three methods for determining the DNA concentration of the genomic DNA extracts from maize events Bt11, Mon810 and NK603 were compared. DNA extracts were derived from a similar quantity of starting material (40 milligrams as defined by the extraction protocol with multiple extracts combined to exceed the 100 milligrams ERM minimum recommendation) and extracted to the same protocol. Left: NanoDrop spectrophotometer results for each maize event in duplicate (the 260:280 nanometre ratios for all samples ranged from 1.71 to 1.81 and the ratio 260:230 nanometre from 1.69 to 2.59, Bt11 concentration 114.3 nanograms per microlitre); Centre: duplicate loading of DNA from each maize event on an agarose gel (full size gel image shown in Fig. S9) compared to ladder of defined DNA concentrations showing intact genomics DNA of large fragment size (Bt11 concentration 52.0 nanograms per microlitre); Right the three quantification methods compared (Qubit determined Bt11 concentration 51.4 nanograms per microlitre).