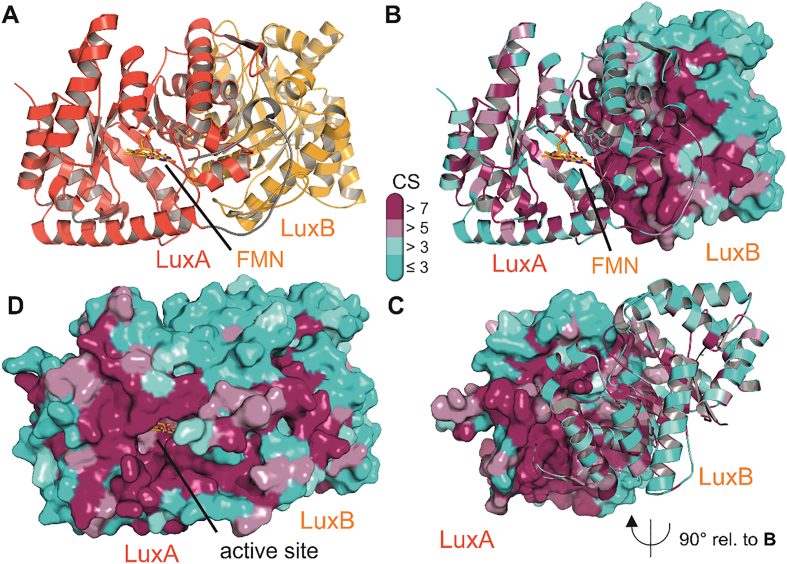
Fig. 3.
Crystal structure of the bacterial luciferase from Vibrio harveyi (PDB 3FGC). Panel A shows the characteristic heterodimer of LuxA (red) and LuxB (orange) in cartoon representation. The FMN cofactor in the active site is shown as yellow stick model. The characteristic loop region that mediates the contact between the α- and β-subunits is shown in grey [15]. Panels B, C and D feature the same luciferase dimer colored according to the conservation of residues among the members of bioluminescent bacteria (details of which sequences are aligned are shown in supplementary Table 1). Conservation scores from 1 to 9 correspond to an increase in evolutionary conservation and are colored according to the bar legend in the middle of the figure with higher scores (purple) indicating higher conservation. Panels B and C show the high conservation of residues at the heterodimer interface. Either LuxB or LuxA are shown in surface representation in panels B and C, respectively, and panel C features a 90° out of plane rotation of the dimer for better visibility of the highly conserved LuxA interface. Panel D shows both protomers in surface representation and highlights the strict conservation of residues in the open active site as well as its entrance. Conservation scores were computed with the ConSurf server [11].