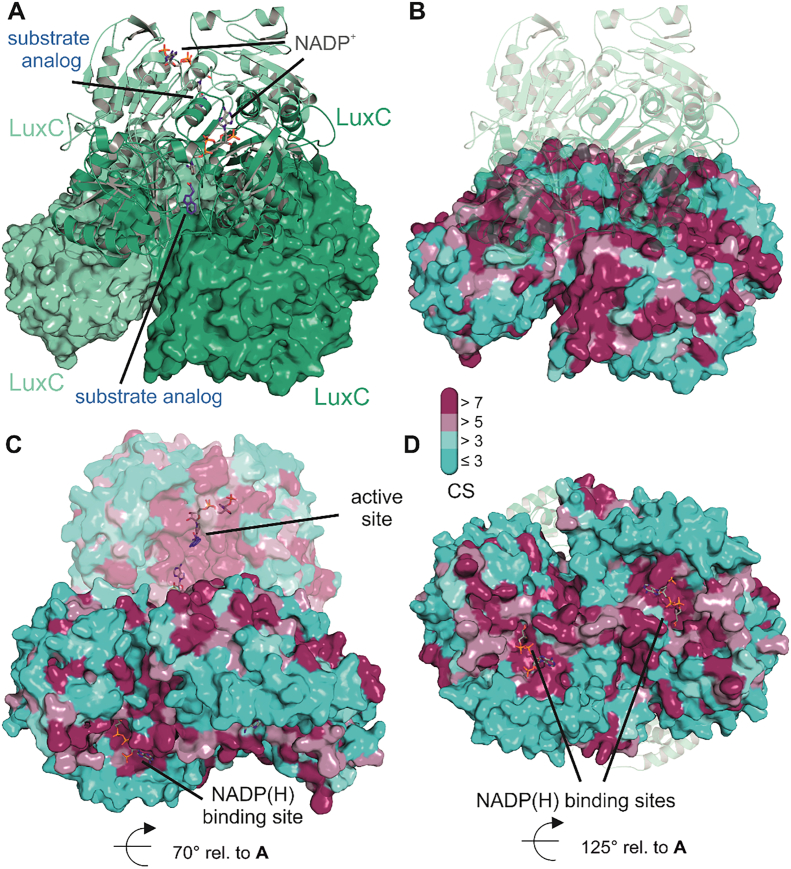
Fig. 6.
Homology model of LuxC from V. harveyi. Panel A features the tetrameric assembly of LuxC obtained from the SWISS-MODEL server [95] using the crystal structure of methylmalonate semialdehyde dehydrogenase from Bacillus subtilis as template (PDB 1T90). The tetramer corresponds to a dimer of dimers, which are shown once as cartoon representation and once as molecular surface. Individual protomers of each dimer are colored in green and light green. For the dimer in cartoon representation, we also show stick models of the substrate analog (indole-3-acetaldehyde – blue) and the cofactor (NADP+ − grey) in the respective binding sites obtained from the superposition of the LuxC model with indole-3-acetaldehyde dehydrogenase from Pseudomonas syringae (PDB 5IUW) and from the structure of an aldehyde dehydrogenase from Burkholderia multivorans (PDB 5JRY), respectively. Panels B, C and D provide an overview of the evolutionary conservation of residues according to the ConSurf server [11] and computed conservation scores (CS – bar legend). Panel B highlights the conserved residues at the interface of the individual LuxC dimers in the same orientation as panel A. For clarity, the dimer in cartoon representation is shown in transparent mode. Panel C shows a different orientation of the tetrameric assembly, to demonstrate the high degree of conservation in the active site generated at the interface of two subdomains of each LuxC protomer. Importantly, the substrate and the NADPH cofactor approach the active site from opposite sides. Panel D shows another view of one LuxC dimer highlighting the conservation of residues around the NADP+ binding site. For better visibility loop regions covering the NADP+ binding site are not shown in this figure. Similar to the observations for LuxE, patches of strongly conserved residues can be found at surface elements near the active site that are not involved in LuxC oligomerization and are therefore likely involved in complex formation with the LuxE synthetase subunits.