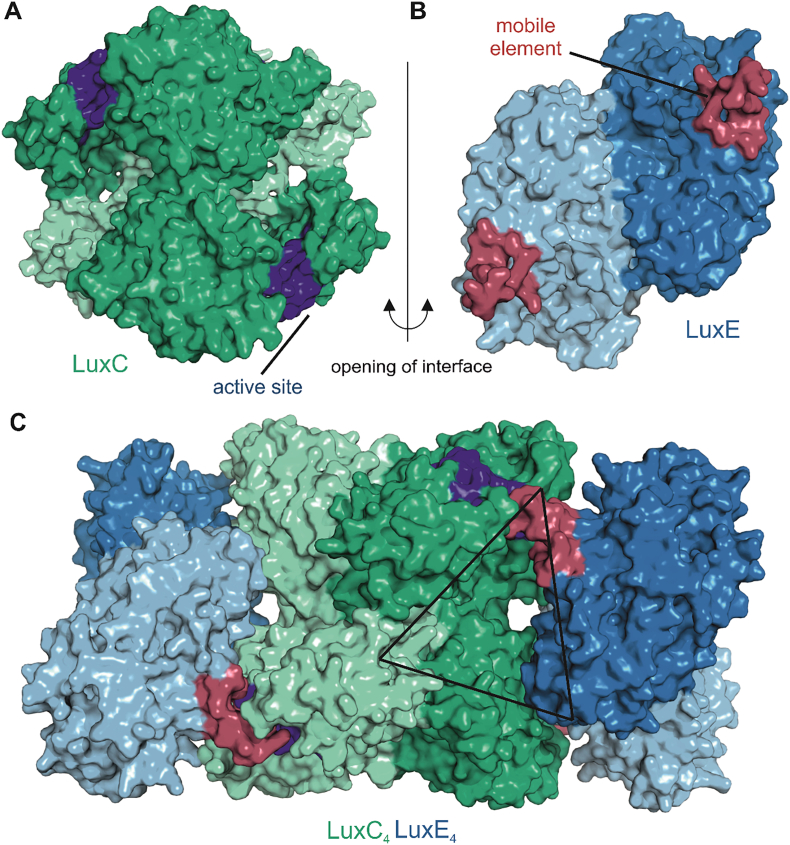
Fig. 7.
Model of the LuxC - LuxE interaction. Panels A and B show surface representations of LuxC and LuxE, respectively, colored according to the details presented in Fig. 5, Fig. 6. The active site region of LuxC is highlighted in dark blue and the mobile element of LuxE containing the acylated residue is colored in red. The central region of complementarity at the LuxC2-LuxE2 interface is lined by many highly conserved residues at specific surfaces of the respective oligomeric structures that have been highlighted in Fig. 5D (LuxE) and Fig. 6B (LuxC). Their complementarity is illustrated by the opening of the interface (right side interface of the complex in panel C) by opposite 90° rotations of LuxC and LuxE. Panel C also shows the second LuxE dimer bound to the opposite side of the LuxC tetramer resulting in an overall LuxC4LuxE4 stoichiometry. The architecture around the active site reveals an interesting triangular complementarity to the LuxD structure (Fig. 4). For generation of the LuxC-LuxE complex we used a different template for homology modeling of LuxC to better reflect the open apo-conformation that might be needed for the initial interaction with LuxE. This model was again generated with the SWISS-MODEL server [95] based on the apo form of the indole-3-acetaldehyde dehydrogenase (PDB 5IUU). The apo form is characterized by a modest opening of the active site accompanied by unstructured loop regions involved in substrate coordination.