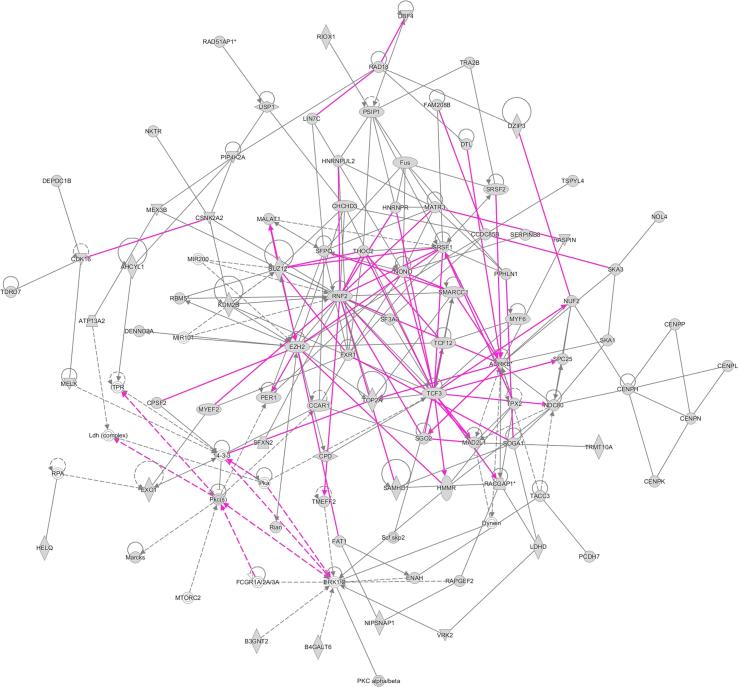
Fig. 3.
Molecular network generated using Ingenuity Pathway Analysis (IPA) software. The merged network was generated from the top three networks expressed in a list of 513 probe sets (representing 443 different genes) differentially expressed in non-diabetic and T1D mice. The list of genes was generated from T test unpaired unequal variance (p < 0.005, no multiple test correction) of mRNA expression data whose expression was altered in T1D mice compared to non-diabetic mice. Genes from our uploaded gene list (focus genes) are shown as gray icons while genes or endogenous chemicals derived from the IPA database that could be algorithmically connected to these focus genes are shown as white icons. The shapes of the icons represent classes of genes/molecules e.g. squares = cytokines/growth factors, ovals = transcription factors, etc.