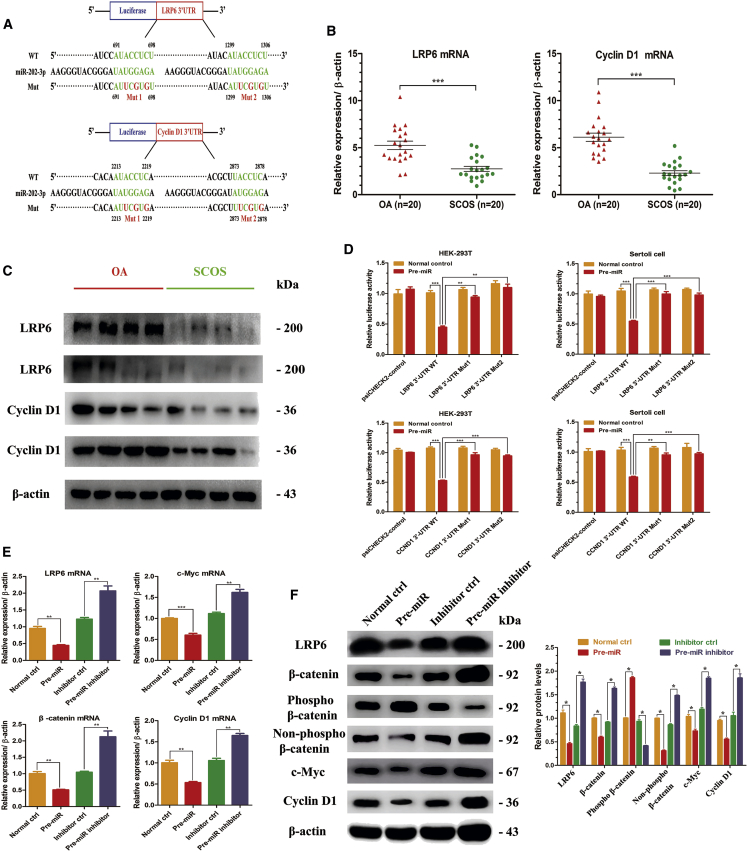
Figure 5.
Identification of Direct Targets of miR-202-3p in Human Sertoli Cells
(A) There were two potential binding sites of miR-02-3p at the 3′ UTR region of LRP6 and Cyclin D1 mRNA based on the bioinformatic analysis. 3′ UTR of LRP6 and Cyclin D1 mRNA containing wild-type (WT), mutant-1 (Mut1), and mutant-2 (Mut2) was cloned into dual-luciferase plasmids. (B) The mRNA levels of LRP6 and Cyclin D1 were detected using real-time qPCR in 20 paired OA and SCOS Sertoli cells. (C) Western blots revealed the different expression patterns of LRP6 and Cyclin D1 between OA and SCOS Sertoli cells (n = 8). β-actin served as a loading control. (D) Empty plasmids (psiCHECK2 control) and dual-luciferase plasmids containing WT, Mut1, or Mut2 of LRP6 or Cyclin D1 3′ UTR were transfected into HEK293T cells and OA Sertoli cells with miR-202-3p overexpression plasmids or control plasmids, respectively. Firefly and Renilla luciferase signals were performed for luciferase activity after 36 hr of transfection. (E) mRNA levels of LRP6, c-Myc, Cyclin D1, and β-catenin, as measured by real-time qPCR in the four cell strains. (F) Western blots showed the LRP6, c-Myc, Cyclin D1, β-catenin, phospho-β-catenin, and non-phospho-β-catenin protein levels at 72 hr after virus infection and puromycin screening. β-actin served as a loading control. Results of the pre-miR group were normalized to the normal ctrl group, and results of the pre-miR inhibitor group were normalized to the inhibitor ctrl group. *p < 0.05; **p < 0.01; ***p < 0.001.