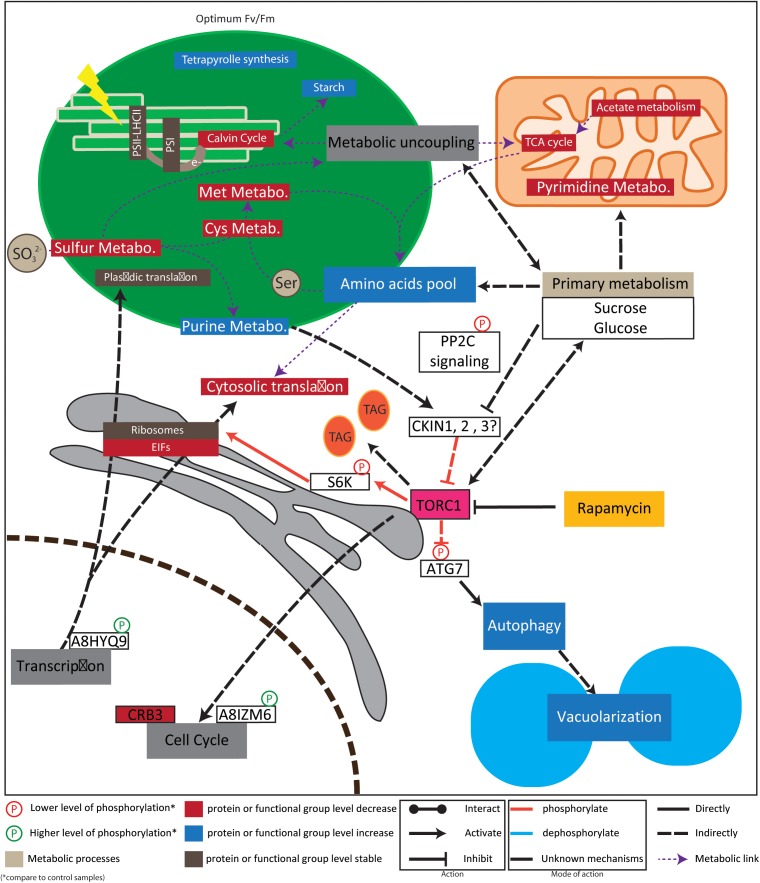
FIGURE 7.
Model for regulation of rapamycin treatment response. Intrestingly, our data suggest that chloroplastic function are not inhibited but rather adapted. Especially, plastid localized, Calvin cycle, sulfur, cystein and methionine related proteins are down regulated. Instead, starch accumulate and purine metabolsime is stimulated. Those observation are line with previous reports indicating a connection of sulfur metabolism with TOR signaling (Dong et al., 2017). Together with our results, recent metabolic data suggest also a metabolic uncoupling between the plastid and mitochondria, associate with a decrease of mitochondrial metabolic activity (Juppner et al., 2017). In line with plastid maintenance, ribosomes didn’t present clear regulation pattern, instead EIF factors were down regulated. Reduced cytosolic translation activity is in line with S6K phosphorylation previously shown to be responsible for translation regulation (Dobrenel et al., 2016b). This reduced cytosolic translation activity could explain amino acid accumulation previously detected (Juppner et al., 2017). Further, protein associated to circadian cycle and flowering time regulation were together decreasing in protein abundance or phosphorylation level. Finally, phosphorylation of ATG7 is in line with previously detected autophagy induction and cell vacuolarization (Crespo et al., 2005; Couso et al., 2017). For example, the red- arrow between TOR and SK6 indicate that TOR complex directly and positively regulates RPS6 by phosphorylation. PS is photosystem; LHCII is Light-Harvesting complex-II; e- represents electron from the electron transfer chain; TAG stands for tri-acy-glycerol; Cys for cysteine; Met for methionine; for protein accession number and name refer to results part.