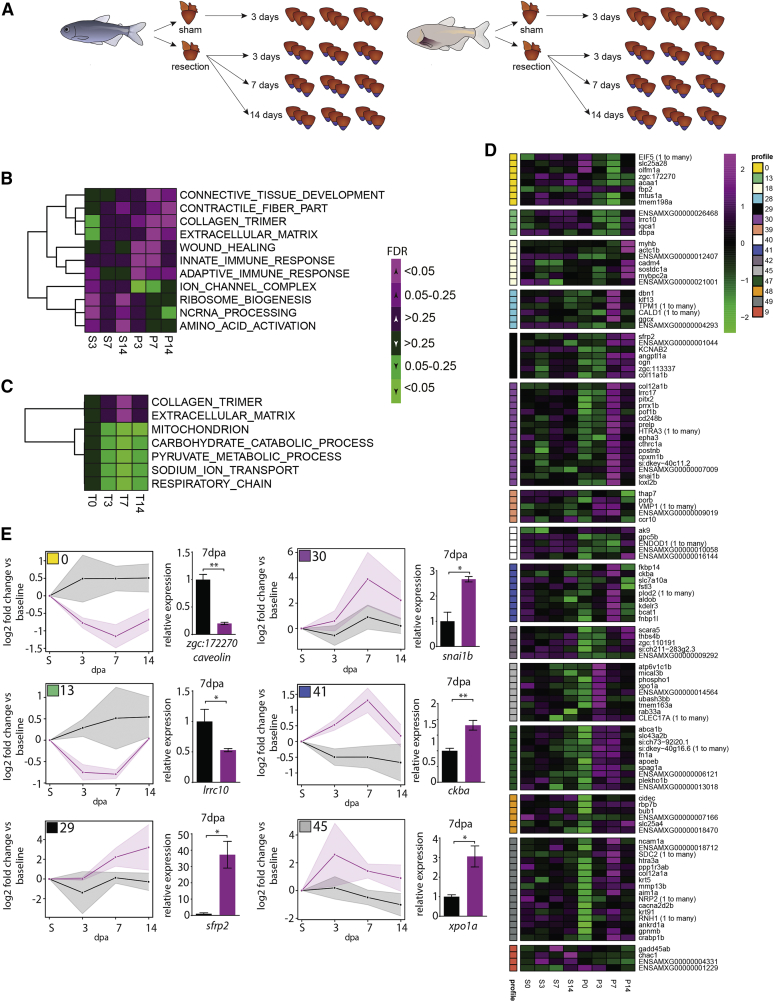
Figure 3.
Analysis of the Kinetics of Differential Expression in Response to Wounding Identifying lrrc10
(A) Experimental design. Three ventricles pooled per sample, three samples per time point.
(B) Heatmap of signed false discovery rate (FDR) values for selected Gene Ontology (GO) terms significantly different by pre-ranked gene set enrichment analysis (GSEA; FDR < 0.05) between sham and at least at one time point per fish.
(C) Heatmap of signed FDR values for selected GO terms significantly different between Pachón and surface fish per time point (pre-ranked GSEA FDR < 0.05 at least at one time point).
(D) Heatmap of row-normalized gene expression with differential kinetics over the different time points grouped by profile. Profiles shown only if containing at least four genes.
(E) Graphs showing median differences in kinetics of selected profiles (profile numbers and colors based on C) between surface fish (gray) and Pachón (magenta), as well as qPCR validation of one of the genes in the same profiles at 7 dpa. Shaded areas show median absolute deviation. For qPCRs, n = 3 for both Pachón and surface fish, unpaired two-tailed t test.
Detailed statistics are provided in STAR Methods. Results are presented as mean ± SEM.