Fig. 5.
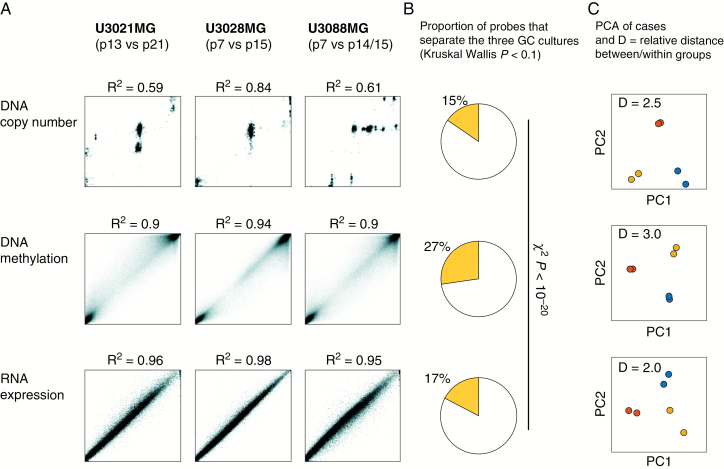
DNA methylation is stable in glioblastoma cell cultures. We analyzed the effect of clonal takeover events on each of (i) DNA copy number, (ii) DNA methylation and (iii) RNA transcription. (A) Scatter plots of early vs late time points; while there was a numerical change in DNA copy number, DNA methylation and RNA transcription remained more stable. (B) Each measured gene locus, transcript, and methylation event was scored by Kruskal‒Wallis test to identify observations that were significantly different between U3021MG, U3028MG, and U3088MG, viewing the early and late passage time points as within-group replicates. DNA methylation had a higher fraction of observed variables with Kruskal‒Wallis P-value < 0.1. The significance of the altered fraction was confirmed by the chi-square test (P < 10−20). (C) PCA, indicating separation of the 3 GC cultures, showing the 2 time points of each GC in the same color. While the PCA plot is different for each platform, the relative average distance between the GC cultures compared with the average distance within the GC cultures was higher for DNA methylation profiles (3.0 vs 2.0–2.5).