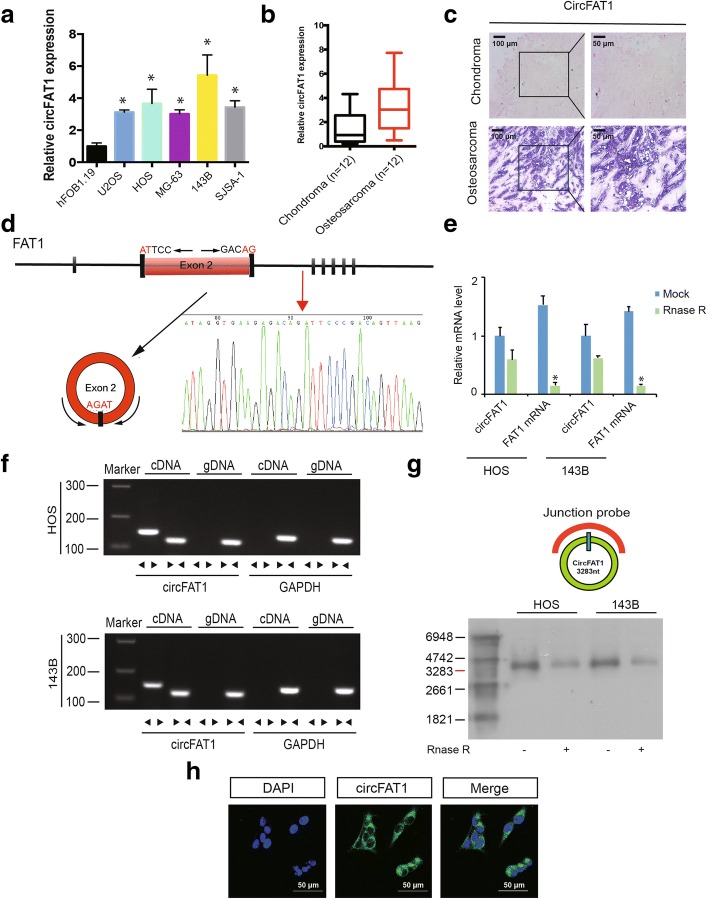
Fig. 1.
CircFAT1 validation and expression in osteosarcoma tissue and cells. a CircFAT1 expression in hFOB1.19 and osteosarcoma (OS) cell lines (OS-732, 143B, HOS, SJSA-1, and MG-63) was evaluated by qRT-PCR. Data represents the mean ± standard deviation (SD) (n = 3). * P < 0.05 (b) circFAT1 expression was higher in human OS than in chondroma tissue. Data represents the mean ± SD (n = 12). c CircFAT1 expression was higher in human OS than in chondroma tissue. Representative images are shown (400× magnification). d Schematic illustration showed the FAT1 exon 2 circularization forming circFAT1 (black arrow). The presence of circFAT1 was validated by RT–PCR, followed by Sanger sequencing. Red arrow represents “head-to-tail” circFAT1 splicing sites. e The expression of circFAT1 and FAT1 mRNA in HOS and 143B cells treated with or without RNase R was detected by real-time PCR. The relative levels of circFAT1 and FAT1 mRNA were normalized to the value measured in the mock treatment. Data represents the mean ± SD (n = 3). * P < 0.05. f The presence of circFAT1 was validated in HOS and 143B osteosarcoma cell lines by RT–PCR. Divergent primers amplified circFAT1 in cDNA, but not in genomic DNA. GAPDH was used as a negative control. g Northern blots for detecting circFAT1 in HOS and 143B cells treated with or without RNase R digestion. The upper panels show the probed blots of circFAT1, and the red triangle represents circFAT1 band size (3283 base pairs (bp)). The lower panels show the gel electrophoretic results of RNA with or without RNase R digestion. h RNA fluorescence in situ hybridization (FISH) showed that circFAT1 was predominantly localized in the cytoplasm. CircFAT1 probes were labeled with Alexa Fluor 488, Nuclei were stained with DAPI. Scale bar, 50 μm