Abstract
The development of gastric cancer is frequently related to the overexpression of wild-type p21 proteins, but it is rarely related to mutated Ras proteins. We previously constructed a broad-spectrum anti-p21-Ras single-chain variable fragment antibody (scFv), which was carried by the oncolytic adenovirus KGHV500. Here we explored the antitumor effects of this recombinant oncolytic adenovirus carried by cytokine-induced killer (CIK) cells on human gastric SGC7901 cells that overexpress wild-type Ras. The MTT assay, scratch test, Transwell assay, and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay were performed in vitro to investigate the proliferation, migration, invasiveness, and cell apoptosis rate, respectively, of the human gastric cell line SGC7901 treated with KGHV500 adenovirus. Then, the tumor-targeting ability and systemic safety of KGHV500 adenovirus delivered by CIK cells were explored in vivo. We found that KGHV500 adenovirus could significantly inhibit proliferation, migration, and invasiveness and promote cell apoptosis in SGC7901 cells in vitro. In vivo studies showed that CIK cells could successfully deliver KGHV500 adenovirus to the tumor site; the two vectors synergistically killed tumor cells, and the treatment was relatively safe for normal tissues. In conclusion, this therapeutic strategy of recombinant adenovirus KGHV500 delivered by CIK cells offers a positive prospect for the targeted therapy of Ras-related cancers.
Keywords: oncolytic adenovirus, anti-p21-Ras, scFv, CIK cell, gastric cancer, SGC7901
Introduction
Gastric cancer is the fourth most common cancer worldwide and the second leading cause of cancer-related mortality in humans.1 Current therapies for gastric cancer are traditional surgical treatment and chemotherapy. Studies showed that adjuvant chemotherapy produced a 6% increase in the 5-year overall survival of resectable gastric cancer patients.2, 3 However, chemotherapy is not effective in patients diagnosed at an advanced or unresectable stage. Molecularly targeted therapies provide opportunities for gastric cancer patients with poor prognosis. Trastuzumab is a currently approved targeting agent for the treatment of HER2-positive gastric cancers, and it increases the median overall survival of patients up to 13.8 months, compared with the 11.1-month improvement in overall survival produced by chemotherapy alone.4 The rate of HER2 overexpression in gastric cancer is approximately 22%,5 which indicates that only a few subsets of patients could receive benefits from trastuzumab. A large number of HER2-negative gastric cancer patients could not receive any benefit. As it is known that the development of gastric cancer is a complex and multistep process, gastric cancer treatment will not be a simple one-size-fits-all therapy. Therefore, searching for new attractive candidate targets and developing relevant therapy agents are urgent needs in gastric cancer therapy.
Ras genes, classic proto-oncogenes, play an important role in cell growth and differentiation. Abnormally activated Ras proteins act as molecular switches in the processes of malignant transformation and tumorigenesis.6 Disappointingly, clinically effective anti-p21-Ras therapies have remained elusive until now, and Ras proteins have even been considered “undruggable.”7, 8, 9 We developed an anti-p21-Ras single-chain variable fragment antibody (scFv) in previous studies, and we found that it could bind with all the wild-type and partially mutated H-Ras, K-Ras, and N-Ras proteins that exist in different human tumor cell lines and primary tumor tissues.10 Then, this anti-p21-Ras scFv gene was cloned into adenovirus vectors, and the recombinant vectors showed obvious antitumor effects on several tumor cell lines in vivo and in vitro.11
To decrease the infection of normal cells, the adenovirus vectors were modified to be oncolytic viral vectors, where the endogenous E1a and E1b promoters of the adenovirus were replaced with the human telomerase reverse transcriptase (hTERT) and hypoxia response element (HRE) promoters, two tumor-specific promoters. This engineered adenovirus was called recombinant oncolytic adenovirus KGHV300, and it exhibited significant antitumor activity in several tumors in previous studies.12 However, it could only be administered by intratumoral injection, because it can infect normal cells even though it cannot replicate in normal cells.
To increase the tumor cell-targeting ability and systemic administration safety of the KGHV300 adenovirus, its cilia gene was modified so that it could bind to cytokine-induced killer (CIK) cells, and, thus, the new recombinant oncolytic adenovirus KGHV500 was constructed (unpublished data). It is well known that CIK cells have potent intratumoral homing ability in gene therapy,13 and they are often used as effective pharmacological tools for cancer immunotherapy.14 In this study, we presumed that CIK cells could be used as a targeted delivery vehicle for KGHV500 adenovirus in vivo, which could largely decrease the infection of normal cells. We investigated the effects of anti-p21-Ras scFv delivered by KGHV500 adenovirus and CIK cells on human gastric cancer SGC7901 tumor xenograft models. Furthermore, the effects of KGHV500 adenovirus on SGC7901 cell proliferation, migration, and invasiveness were observed, as well as its effects on cell apoptosis and related genes.
Results
Recombinant Oncolytic Adenovirus KGHV500 Infected SGC7901 Cells
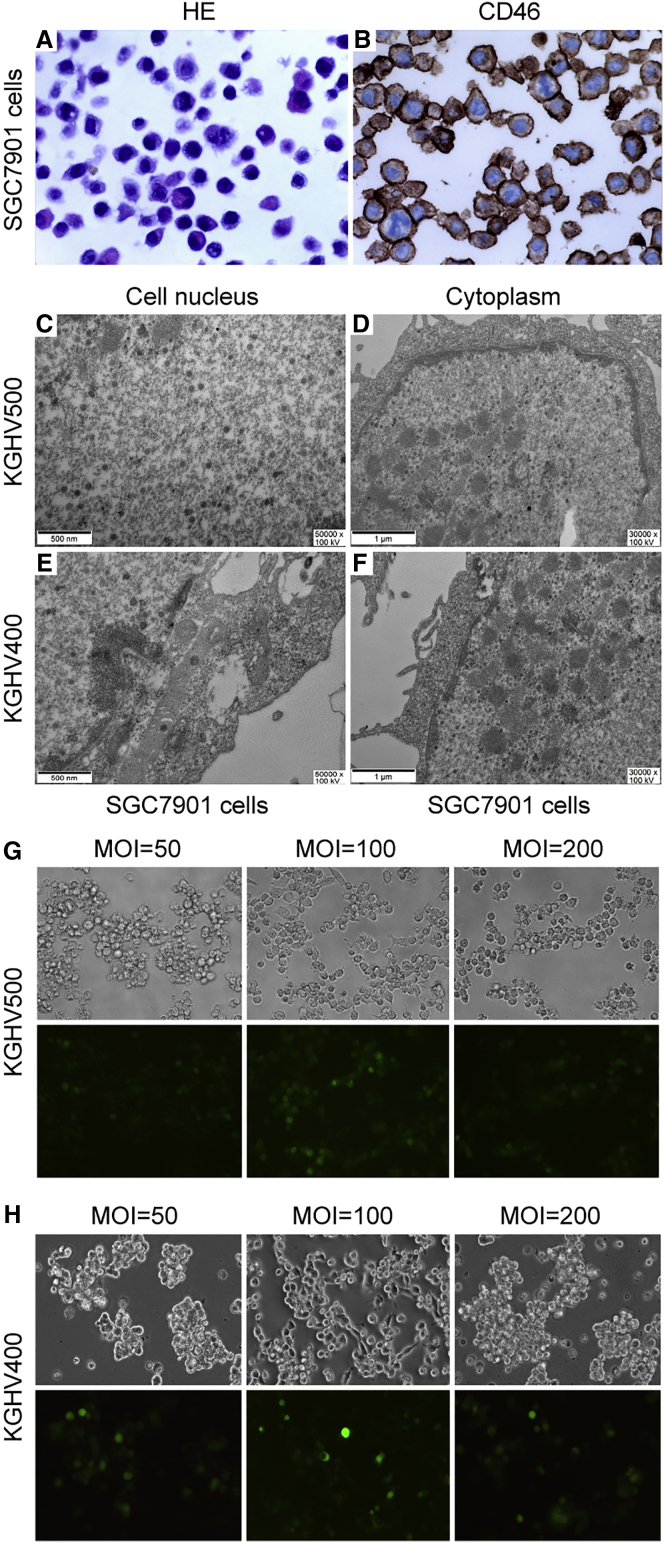
SGC7901 cells that were viewed by microcopy exhibited the following significant atypia: larger cell size, larger and deeply dyed cell nucleus, and higher nucleoplasmic ratio (Figure 1A). Because KGHV500 and KGHV400 adenoviruses infected cells by binding to CD46 molecules, the receptor of adenovirus cilia protein, on the cell surface, we detected the expression of CD46 receptors on SGC7901 cells by immunohistochemistry, and we found that the percentage of positive cells was nearly 100% (Figure 1B). These results revealed that KGHV500 and KGHV400 adenoviruses could infect SGC7901 cells. The description of these adenoviruses and the differences between them are detailed in Figure S1.
Figure 1.
Recombinant Oncolytic Adenovirus KGHV500 Infected SGC7901 Cells
(A) SGC7901 cells were collected, fixed, dehydrated, paraffin embedded, sectioned, stained for H&E, and observed with a microscope at 400×. The cells exhibited the following significant atypia compared to normal cells: cell size varied, cell nuclei were larger and deep dyeing, and the nucleoplasmic ratio was higher. (B) The paraffin-embedded SGC7901 cells were sectioned, and anti-CD46 mAb was used to detect whether CD46 protein was expressed on SGC7901 cell surfaces. It was found that nearly all the cells expressed CD46 protein, which was the receptor for the recombinant oncolytic adenoviruses KGHV500 and KGHV400. (C–F) To further identify whether SGC7901 cells were infected by KGHV500 and KGHV400 adenoviruses, the cells were cocultured with KGHV500 or KGHV400 adenovirus at an MOI of 100 for approximately 48 hr, then observed by transmission electron microscopy. KGHV500 adenovirus particles were found in the SGC7901 cell nucleus (C) and cytoplasm (D), and KGHV400 adenovirus particles were found in the SGC7901 cell nucleus (E) and cytoplasm (F). (G) The infection efficiency of KGHV500 adenovirus on SGC7901 cells was detected by TCID50, and an MOI of 100 was the best infection efficiency. (H) An MOI of 100 was also the best infection efficiency for KGHV400 adenovirus on SGC7901 cells.
To further identify whether KGHV500 and KGHV400 adenoviruses infected SGC7901 cells, transmission electron microscopy was used to observe adenovirus particles in SGC7901 cells after infection for 24 hr. KGHV500 adenovirus particles (70–90 nm) were found in the cell nucleus and cytoplasm of the SGC7901 cells (Figures 1C and 1D), and KGHV400 adenovirus particles were also found in SGC7901 cells (Figures 1E and 1F).
Furthermore, the highest infection efficiencies for KGHV500 and KGHV400 adenoviruses on SGC7901 cells were detected by 50% tissue culture infective dose (TCID50) assay. SGC7901 cells were treated with KGHV500 or KGHV400 adenovirus at an MOI of 10, 25, 50, 100, or 200. Then 48 hr later, the infection efficiency was observed with an inverted fluorescence microscope. The results showed that the fluorescence signal was weak at MOIs of 10, 25, and 50, but the signal increased up to 80% at an MOI of 100, and the cells showed a weak cytopathic effect (CPE) phenomenon compared with cells treated with an MOI of 200, which showed a low fluorescent signal with an excessively strong CPE phenomenon. Therefore, the MOI of 100 yielded the best infection efficiency for both KGHV500 (Figure 1G) and KGHV400 (Figure 1H) adenoviruses.
Inhibitory Effects of KGHV500 Adenovirus on SGC7901 Cells In Vitro
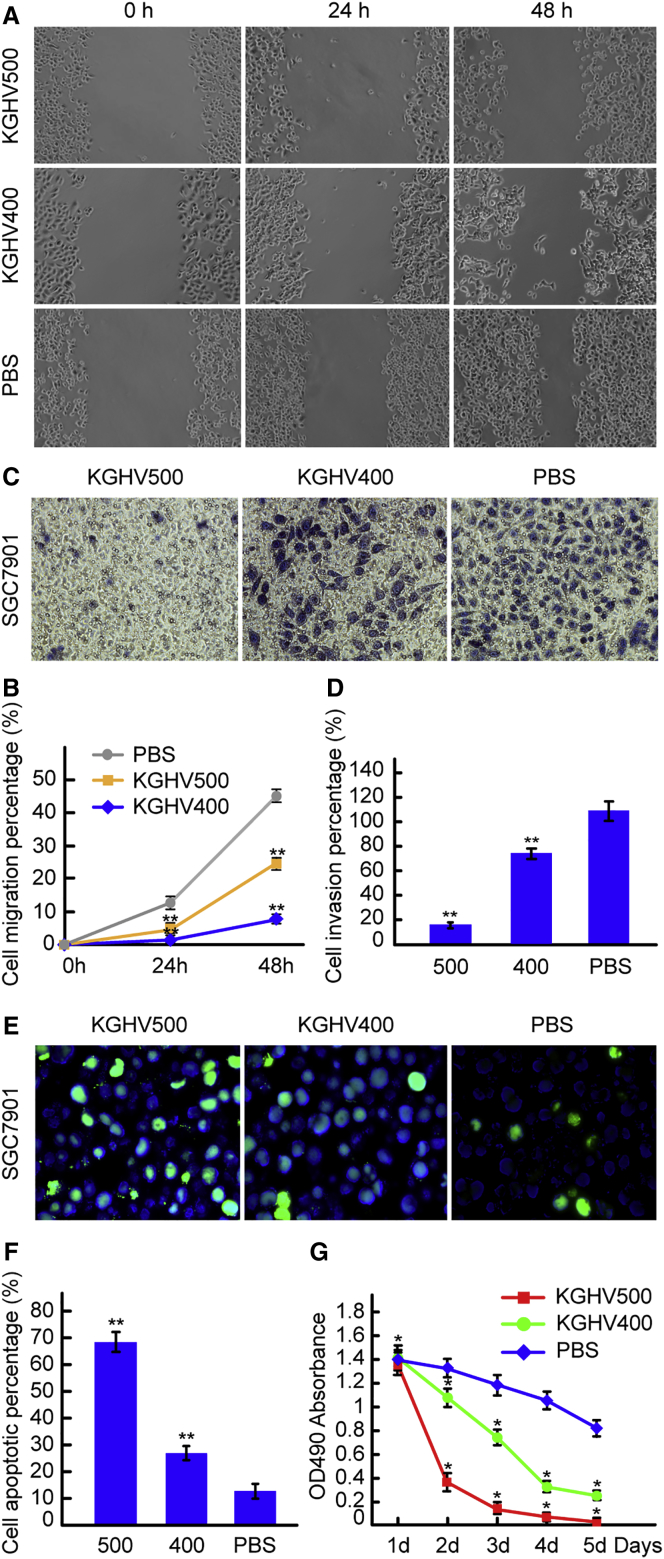
To investigate the effect of KGHV500 adenovirus on SGC7901 cell migration ability, a scratch test was conducted. The scratch wounds of the KGHV500 group showed slower healing than those of the KGHV400 group and PBS group 24 and 48 hr after scratching (Figure 2A). The percentages of cell migration in the KGHV500 group were 1.89% ± 0.29% at 24 hr and 8.67% ± 0.55% at 48 hr; in the KGHV400 group, the percentages of cell migration were 3.85% ± 0.19% at 24 hr and 18.20% ± 0.86% at 48 hr, and, in the PBS group, the percentages of cell migration were 8.25% ± 0.46% at 24 hr and 22.06% ± 1.13% at 48 hr (Figure 2B). There was a significant difference between the KGHV500 group and the other two groups (p < 0.01). This result indicated that recombinant oncolytic adenovirus KGHV500 could inhibit the migration ability of SGC7901 cells.
Figure 2.
Effects of KGHV500 Adenovirus on SGC7901 Cells In Vitro
(A) A scratch test was employed to investigate the migration ability of SGC7901 cells treated with KGHV500 adenovirus. The scratch wound of the KGHV500 group showed slower healing than the scratch wounds of the KGHV400 and PBS groups at 24 and 48 hr. (B) The percentage of migrating cells was calculated, and, compared with the KGHV400 and PBS groups, in the KGHV500 group it was lower at 24 and 48 hr (mean ± SEM; **p < 0.01, one-way ANOVA). (C) A Transwell assay was used to detect the effects of KGHV500 adenovirus on the invasiveness of SGC7901 cells. The invasive cells were investigated under microscopy at 200× magnification. Only a few cells invaded into the lower layer in the KGHV500 group, as compared with the KGHV400 and PBS groups. (D) The number of invasive cells was counted, and in KGHV500 group it was lower than in the KGHV400 and PBS groups (mean ± SEM; **p < 0.01). (E) The apoptosis of SGC7901 cells treated with KGHV500 adenovirus was investigated by TUNEL assay, and the apoptotic cells were observed with a fluorescence microscope at a magnification of 1,000×. The number of apoptotic cells in the KGHV500 group was higher than in the KGHV400 and PBS groups. (F) The percentage of apoptotic SGC7901 cells in the KGHV500 group was higher than in the KGHV400 and PBS groups (mean ± SEM; **p < 0.01). (G) The effect of KGHV500 adenovirus on SGC7901 cell proliferation was observed by MTT assay. The OD490 absorbance of SGC7901 cells in the KGHV500 group was significantly lower than in the KGHV400 and PBS groups (mean ± SEM; *p < 0.05).
Transwell chamber invasion test results showed that the number of KGHV500-infected SGC7901 cells that invaded into the lower layer was significantly reduced compared to the number that invaded in the KGHV400 and PBS groups (Figure 2C). The number of invasive cells in the KGHV500 group was 15.0 ± 2.94; however, in the KGHV400 group it was 72.0 ± 4.03, and in the PBS group it increased to 108.57 ± 8.19 (Figure 2D). The differences among the three groups were significant (p < 0.01).
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assays were employed to detect the effects of KGHV500 adenovirus on the apoptosis of SGC7901 cells. The number of apoptotic cells in the KGHV500 group was distinctly increased, whereas the numbers of apoptotic cells in the KGHV400 adenovirus and PBS groups were lower (Figure 2E). The percentage of cells undergoing apoptosis was 68.36% ± 3.95% in the KGHV500 group, but it was 26.60% ± 2.92% in the KGHV400 group and 12.62% ± 2.77% in the PBS group (Figure 2F). There were significant differences among the above groups (p < 0.01), which indicated that the recombinant oncolytic adenovirus KGHV500 could promote the apoptosis of SGC7901 gastric cancer cells.
An MTT assay was carried out to investigate whether the KGHV500 adenovirus could inhibit the proliferation of SGC7901 cells. The cell viability of KGHV500-infected SGC7901 cells decreased significantly compared to the cell viability in the KGHV400 group and PBS group. The absorbance values in the KGHV500 group at 1, 2, 3, 4, and 5 days were 1.37 ± 0.32, 0.36 ± 0.15, 0.14 ± 0.06, 0.08 ± 0.03, and 0.03 ± 0.01, respectively. However, in the KGHV400 group, the absorbance values were 1.41 ± 0.22, 1.08 ± 0.18, 0.75 ± 0.25, 0.33 ± 0.09, and 0.25 ± 0.13, respectively, and, in the PBS group, they were 1.39 ± 0.43, 1.32 ± 0.37, 1.19 ± 0.29, 1.05 ± 0.21, and 0.82 ± 0.15, respectively (Figure 2G). There were significant differences among the above groups (p < 0.05). This result indicated that the recombinant oncolytic adenovirus KGHV500 could obviously inhibit the proliferation of SGC7901 cells.
CIK Cell Culture, Identification, and Adenovirus-Binding Ability Detection
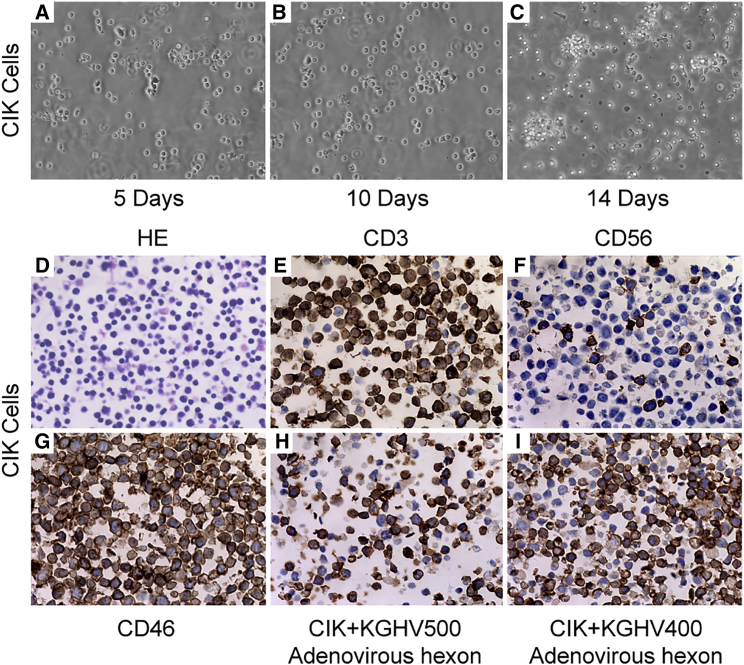
Mononuclear cells were isolated from 20 mL peripheral blood collected from healthy volunteers, and then they were differentiated into CIK cells by the cytokines interferon (IFN)-γ, CD3, interleukin (IL)-1α, and IL-2. The initial CIK cells exhibited suspended growth and remained at the same cell size; 5 days later, the CIK cells gradually formed a cell colony, the cells appeared morphologically irregular, and the number of cells increased rapidly until peaking on the 14th day (Figures 3A–3C). The number of CIK cells increased from 0.5 × 107 to 1.0 × 108 after 14 days, approximately 20× amplification. The CIK cells were embedded into cell wax blocks and viewed by microscopy, which showed that cell sizes were uniform with larger nuclei and less cytoplasm (Figure 3D). Immunohistochemical staining showed that the CIK cell markers CD3 and CD56 were expressed on the cell membrane (Figures 3E and 3F), and the adenovirus receptor CD46 was also expressed on the surface of nearly all the cells (Figure 3G). When CIK cells were cocultured with KGHV500 and KGHV400 adenoviruses at an MOI of 100 for 48 hr, positive staining of the adenovirus hexon was observed on the CIK cell membrane (Figures 3H and 3I), which indicated that KGHV500 and KGHV400 adenoviruses successfully bound to CIK cells.
Figure 3.
CIK Cell Culture, Identification, and Adenovirus-Binding Ability Detection
(A–C) CIK cells were isolated from human peripheral blood and differentiated by adding IFN-γ, anti-CD3 mAb, IL-1α, and IL-2 in different culture stages. CIK cells were observed under a phase contrast microscope at 200× after culturing for 5 (A), 10 (B), and 14 (C) days. Microscope observation showed that the cell number increased gradually and reached its peak and formed a cell colony on day 14, which is a defining characteristic of CIK cells. (D–I) CIK cells were harvested on day 14 and embedded in wax blocks. (D) CIK cells were sectioned and stained with H&E, and they were observed under a microscope at 400× magnification. Microscope observation showed morphologically regular cells, with larger nuclei and less cytoplasm. (E and F) CIK cell markers (E, CD3 and F, CD56) were detected by immunohistochemistry and found to be expressed on CIK cell membranes at 400× magnification. (G) CD46 protein (the receptor for KGHV500 and KGHV400 adenoviruses) was also detected on CIK cell surfaces at 400× magnification. (H and I) KGHV500 (H) and KGHV400 (I) adenovirus hexon was detected on CIK cell membranes at 400× magnification.
Antitumor Effects of CIK Cell-Carried KGHV500 Adenovirus In Vivo
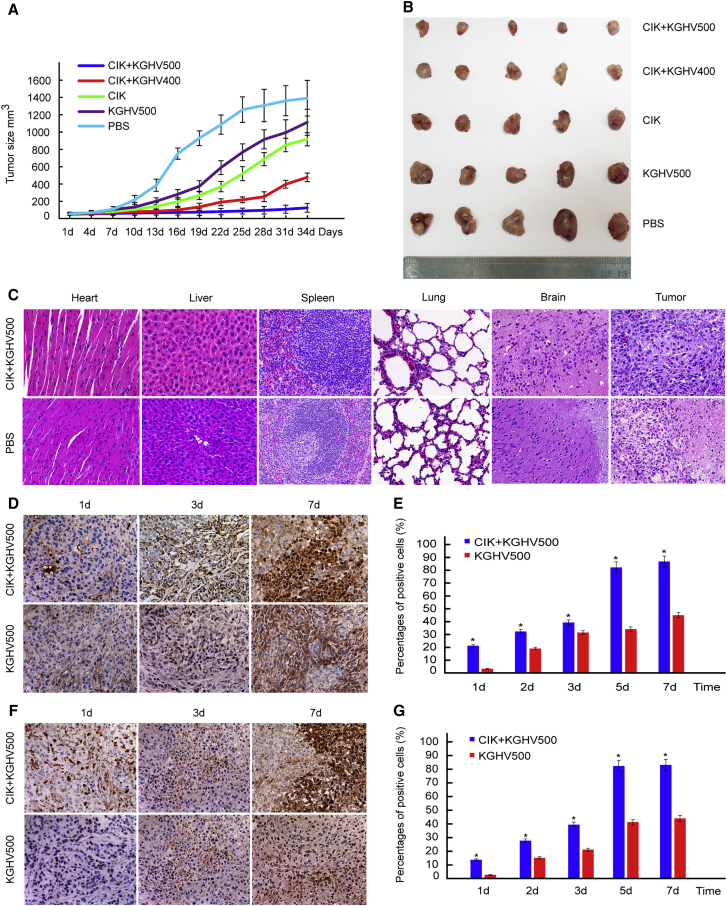
The average tumor diameter reached 5 mm in BALB/c nude mice approximately 3 weeks after subcutaneous inoculation of SGC7901 cells. All the mice were weighed and randomly divided into the following five groups: CIK + KGHV500 group, CIK + KGHV400 group, CIK group, KGHV500 group, and PBS group. The above treatments were administered via tail vein injection. Because some of the mouse tumors grew rapidly and broke through the surface of the skin, presenting as ulceration and bleeding, 34 days after the above treatment, all of the mice were euthanized at that time point. The tumor-bearing nude mice of each group are shown in Figure S2. The tumor growth curves were plotted after giving the different treatments for 34 days (Figure 4A), and the tumor volume from smallest to largest was as follows: CIK + KGHV500 group, CIK + KGHV400 group, CIK group, KGHV500 group, and PBS group (Figure 4B). The comparison among all groups was statistically significant (p < 0.05). CIK cells carrying the recombinant oncolytic adenovirus KGHV500 had a significant inhibitory effect on tumor growth in vivo.
Figure 4.
Antitumor Effects of CIK Cells Carrying KGHV500 Adenovirus In Vivo
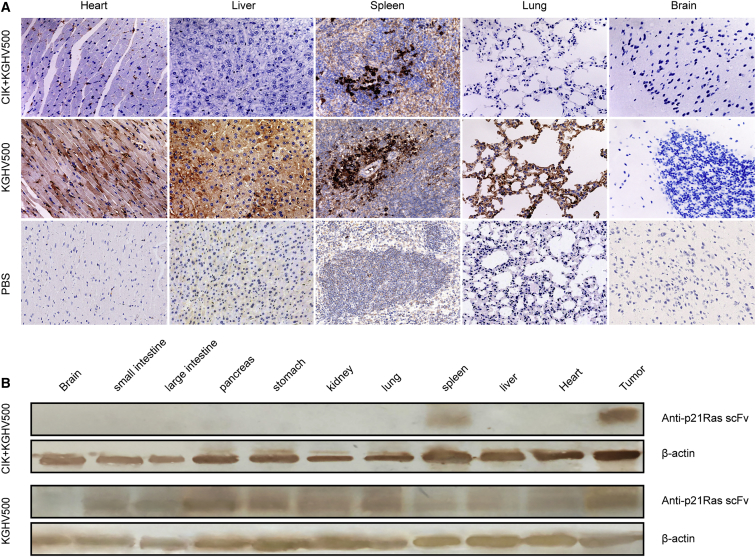
(A) All the tumor-bearing BALB/c-nude mice were weighed and divided into the following five treatment groups: CIK + KGHV500, CIK + KGHV400, CIK, KGHV500, and PBS. These treatments were all performed via tail vein injection. All the mice were euthanized after 34 days due to the tumor tissue in some of the mice ulcerating and bleeding. The tumor growth curves were drawn according to the tumor sizes. They showed that tumor volume from smallest to largest was as follows: CIK + KGHV500, CIK + KGHV400, CIK, KGHV500, and PBS (mean ± SEM; p < 0.05). The increase in tumor volume in the CIK + KGHV500 group exhibited slower progression than the rate of increase in any other group. (B) Tumor tissue from each group was completely dissected and examined by general observation. The results were the same as the above tumor growth curves. (C) All the mouse organs and tumor tissues specimens were sectioned, stained with H&E, and observed with a microscope at 200× magnification. Compared with the PBS group, no obvious abnormalities or lesions were found in the mouse organs, such as the heart, liver, spleen, lungs, and brain, in the KGHV500 group. Tumor tissue from the KGHV500 group, which exhibited a relatively slower growth progression, showed less necrosis and pathological mitosis than tissue from the other groups. (D) KGHV500 adenovirus hexon expressed on tumor cells was observed with a microscope at 200× magnification. The expression of KGHV500 adenovirus hexon reached its highest level on day 7 in the CIK + KGHV500 group and the KGHV500 group. (E) The percentage of positive cells in the CIK + KGHV500 group was much higher than in the KGHV500 group (mean ± SEM; *p < 0.05). (F) The expression of anti-p21-Ras scFv in tumor tissue was also investigated with a microscope at 200× magnification, and it showed the same pattern as the expression of adenovirus hexon. (G) The percentage of cells positive for anti-p21-Ras scFv in the CIK + KGHV500 group was higher than in the KGHV500 group (mean ± SEM; *p < 0.05).
All mouse organs, including the heart, liver, spleen, lungs, kidneys, stomach, pancreas, large intestine, small intestine, and brain, were observed under a microscope. Compared with the PBS group, no obvious abnormalities or lesions were found in the CIK + KGHV500 group (Figure 4C). The tumor tissue from the CIK + KGHV500 group showed less necrosis and pathological mitosis compared with that observed in the tumor tissue from the PBS group, indicating a relatively slower proliferation progress in the CIK + KGHV500 group. All the mouse weights in each group before and after treatment were compared, and none of the differences was statistically significant (p > 0.05) (Table S1), which indicated that the oncolytic adenovirus vector itself did not affect mouse growth.
To investigate the presence of KGHV500 adenovirus in tumor tissue, we detected the expression of adenovirus hexon protein by immunohistochemistry. The expression of adenovirus hexon increased gradually in both the CIK + KGHV500 group and the KGHV500 group, and on day 7 it reached its highest level (Figure 4D). The percentages of adenovirus hexon-positive cells in tumor tissue in the CIK + KGHV500 group were 21.46% ± 3.52%, 32.45% ± 2.79%, 39.51% ± 4.05%, 82.36% ± 8.09%, and 86.95% ± 5.65% on days 1, 2, 3, 5, and 7 after administration, respectively. However, in the KGHV500 group, the percentages were 3.55% ± 1.08%, 19.27% ± 2.64%, 31.65% ± 4.65%, 34.20% ± 1.83%, and 45.19% ± 2.86%, respectively (Figure 4E). The percentages of adenovirus hexon-positive cells were significantly higher in the CIK + KGHV500 group than in the KGHV500 group (p < 0.05).
Moreover, KGHV500 adenovirus inhibits tumor cell growth through the expression of anti-p21-Ras scFv; thus, the expression of anti-p21-Ras scFv in tumor tissues was detected by immunohistochemistry. The FLAG tag of the anti-p21-Ras scFv was stained and detected by immunohistochemistry on days 1, 2, 3, 5, and 7 after administration. The expression of anti-p21-Ras scFv was also increased over time in both the CIK + KGHV500 group and the KGHV500 group, with the highest expression on day 7 (Figure 4F), which was in accordance with the expression of KGHV500 adenovirus hexon. The percentages of positive cells were 14.21% ± 1.67%, 28.04% ± 2.08%, 39.66% ± 4.57%, 82.57% ± 4.69%, and 83.25% ± 4.88% on days 1, 2, 3, 5, and 7, respectively, in the CIK + KGHV500 group; but, in the KGHV500 adenovirus group, the percentages were 3.08% ± 0.44%, 15.74% ± 3.88%, 21.67% ± 3.66%, 41.58% ± 4.3%, and 44.37% ± 3.25%, respectively (Figure 4G). The number of positive cells in the CIK + KGHV500 group was significantly higher than that in the KGHV500 group (p < 0.05).
The distribution of KGHV500 adenovirus in all mouse organs was also detected by immunohistochemistry. Because of the existence of the blood-brain barrier, the adenovirus in both the CIK + KGHV500 group and the KGHV500 group could not penetrate and distribute into the brain. Adenovirus hexon in the CIK + KGHV500 group was not found in any of the organs except the spleen (Figure 5A). The distribution in the spleen was probably related to the lymphocyte-homing characteristics of CIK cells. In contrast, adenovirus hexon was found in all mouse organs in the KGHV500 adenovirus group (Figure 5A). Therefore, the above results indicated that CIK cells could carry KGHV500 adenovirus to the targeted tumor tissue, thereby largely decreasing the distribution of KGHV500 adenovirus into normal organs.
Figure 5.
Distribution of KGHV500 Adenovirus and Anti-p21-Ras scFv in Mouse Organs
(A) The expression of adenovirus hexon protein in all the mouse organs was detected by immunohistochemistry and observed with a microscope at 200× magnification. Adenovirus hexon was not expressed in the brain in either the CIK + KGHV500 group or the KGHV500 group due to the blood-brain barrier. There was no adenovirus hexon distributed in any of the mouse organs except the spleen in the CIK + KGHV500 group, whereas, in the KGHV500 group, adenovirus hexon was found in all the mouse organs, such as the heart, liver, spleen, and lungs. (B) To further confirm the expression of anti-p21-Ras scFv in the tumor and all the mouse organs, western blotting was used to detect it, and this showed that anti-p21-Ras scFv was expressed only in the tumor tissue and spleen in the CIK + KGHV500 group, but it was expressed in all of the mouse organs except the brain in the KGHV500 group.
In addition, the expression of anti-p21-Ras scFv in tumor tissue and all mouse organs was investigated by western blot, which found that anti-p21-Ras scFv was expressed only in the tumor and spleen in the CIK + KGHV500 group; but, in the KGHV500 group, it was expressed in all of the mouse organs except for the brain (Figure 5B). The expression of anti-p21-Ras scFv was in accordance with the distribution of adenovirus hexon, which revealed that the recombinant oncolytic adenovirus KGHV500 was delivered to tumor tissues by CIK cells and continuously expressed anti-p21-Ras scFv.
Tumor Cell Apoptosis In Vivo
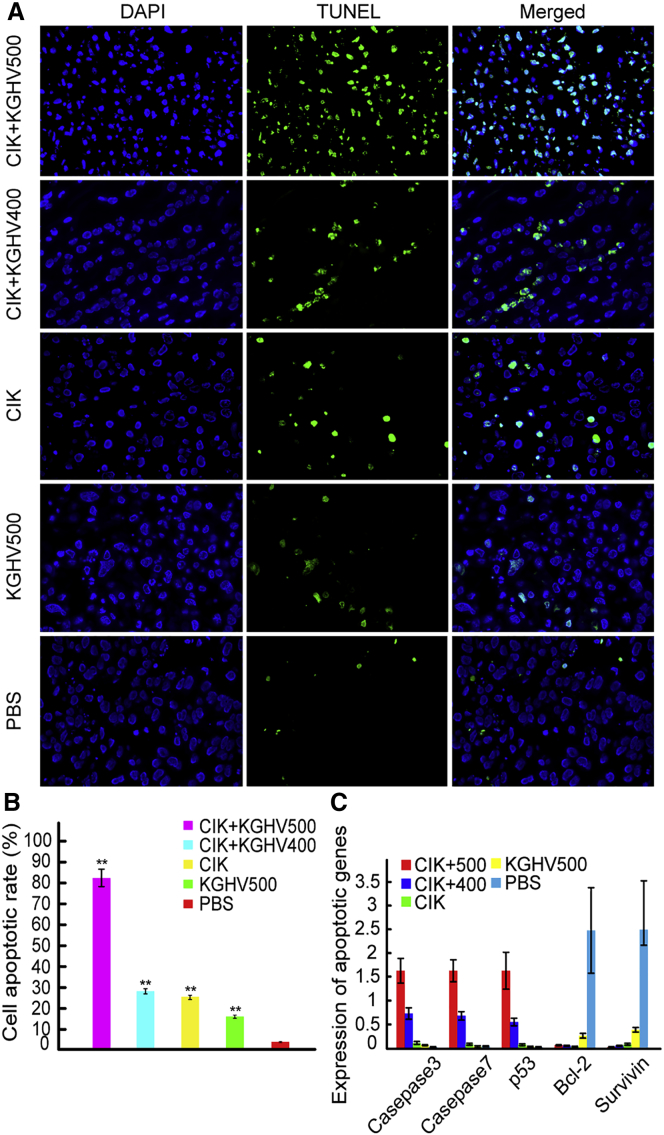
Tumor cell apoptosis was detected by TUNEL assay. Compared with the PBS group, the number of apoptotic cells increased, from lowest to highest as follows: KGHV500 group, CIK group, CIK + KGHV400 group, and CIK + KGHV500 group (Figure 6A). The percentage of apoptotic cells in the CIK + KGHV500 group was 82.68% ± 6.37%, and it was 28.50% ± 5.06% in the CIK + KGHV400 group, 25.72% ± 5.12% in the CIK group, 16.35% ± 2.25% in the KGHV500 group, and 4.56% ± 1.0% in the PBS group (Figure 6B). The comparisons among all the groups were statistically significant (p < 0.01). The recombinant adenovirus KGHV500 carried by CIK cells enhanced the apoptosis of tumor cells in vivo.
Figure 6.
Cell Apoptosis in Tumor Tissues
(A) Tumor tissue blocks were sectioned, and TUNEL assays were performed to investigate the apoptosis of tumor cells. The number of apoptotic cells gradually increased by group as follows: PBS group, KGHV500 group, CIK group, CIK + KGHV400 group, and CIK + KGHV500 group. (B) The percentage of apoptotic cells in the CIK + KGHV500 group was 82.68% ± 11.15%, which is higher than the percentages in the other four groups (mean ± SEM; **p < 0.01). (C) Total RNA was extracted from mouse tumor tissues and visceral tissue, and then the expression of caspase-3, caspase-7, p53, Bcl-2, and survivin genes was detected by qPCR. Compared with the other groups, the expression of pro-apoptotic genes (caspase-3, caspase-7, and p53) increased, but the expression of anti-apoptotic genes (Bcl-2 and survivin) decreased in the CIK + KGHV500 group (mean ± SEM).
Furthermore, the expression of pro-apoptotic genes (caspase-3, caspase-7, and p53) and anti-apoptosis genes (Bcl-2 and survivin) was detected by qPCR in mouse tumor tissue. Compared to the gene expression in the other four groups, the expression of caspase-3, caspase-7, and p53 increased in the CIK + KGHV500 group, but the expression of Bcl-2 and survivin decreased (Figure 6C). Therefore, the KGHV500 adenovirus carried by CIK cells induced tumor cell apoptosis by upregulating the expression of pro-apoptotic genes and simultaneously downregulating the expression of anti-apoptotic genes.
Discussion
Ras oncogenes coding for p21 proteins are frequently involved in the carcinogenesis of various human tumors. For gastric cancer, the total K-Ras mutation frequency is approximately 6%,15 but overexpression of wild-type p21 proteins seems to occur more frequently.16 Studies have indicated that malignant transformation in gastric cancer is rarely associated with activation of Ras genes by point mutations, but it is frequently related to overexpression of wild-type p21 proteins.16, 17 Similarly, our previous studies showed that the overexpression of wild-type p21-Ras proteins, in addition to Ras mutations, may play a prominent role in the development of colorectal cancer and breast cancer and could be a promising target for cancer therapies.18, 19 Therefore, the role of wild-type Ras proteins in human cancers cannot be ignored; in other words, wild-type Ras is also an important therapeutic target in many cancers, including gastric cancer.
In this study, the gastric cancer SGC7901 cell line was used because its three Ras genes are all wild-type, according to the literature.20 Therefore, we aimed to investigate the impacts of anti-p21-Ras scFv carried by the KGHV500 adenovirus on wild-type Ras proteins. The results showed that KGHV500 adenovirus could successfully infect SGC7901 cells and express anti-p21-Ras scFv. The proliferation, migration, and invasiveness of SGC7901 cells were also inhibited by the KGHV500 adenovirus. Moreover, the KGHV500 adenovirus could induce cell apoptosis in SGC7901 cells. The above evidence revealed that this oncolytic adenovirus vector mediated targeted gene therapy that was feasible and effective.
Virus-based gene therapy is the most common and effective approach for cancer gene therapies.21 Its therapeutic efficacy has been widely recognized, resulting in the use of recombinant human p53 adenovirus (Gendicine)22, 23 and T-Vec (talimogene laherparepvec),24, 25 which were approved for clinical cancer treatments in China and the United States, respectively. However, virus-based gene therapy still cannot be systemically administered through the venous injection route to treat deep tumors, such as gastric cancer, because of the inherent risks associated with the viral vector itself. The two main obstacles presently existing in virus-based gene therapies are target specificity and safety of systemic administration.26 To solve these problems, CIK cells were employed to carry the KGHV500 adenovirus to tumor tissue. CIK cells were first reported in 1991 by Schmidt-Wolf et al.,27 who used them to kill human lymphoma cells. CIK cells are a heterogeneous population of effector CD3+ CD56+ natural killer T cells that are artificially differentiated in vitro from peripheral blood mononuclear cells and are commonly used for cancer immunotherapy, as they exhibit major histocompatibility complex (MHC)-unrestricted, safe, and effective antitumor activity.28, 29 In 2006, Thorne et al.29 reported that CIK cells could be used to carry modified vaccinia virus to tumor tissue and synergistically kill tumor cells, which revealed the potential value of this combined therapy in cancer-targeted gene therapy.
In this study, the specific distribution of KGHV500 adenovirus hexon in tumor tissues and the expression of anti-p21-Ras scFv in tumor cells revealed that CIK cells could successfully deliver the recombinant oncolytic adenovirus KGHV500 to the tumor site, which largely decreased the infection of normal tissues. All the internal organs of mice in the CIK + KGHV500 group were carefully examined by pathologists under a microscope, and no obvious abnormalities or lesions were found. The above results demonstrate that CIK cells, as secondary vectors that deliver therapeutic viruses to tumor targets, are effective and safe for systemic administration, and they have the potential to treat deep tumors, such as gastric cancer, through intravenous injection. In conclusion, anti-p21-Ras scFv codelivered by a recombinant oncolytic adenovirus vector and CIK cells showed obvious antitumor activities and targeting ability with relative safety in vivo, which brings optimism to future treatment of the vast majority of Ras-driven tumors, including gastric cancer.
Materials and Methods
Cell Lines and Recombinant Adenovirus
The human gastric cancer cell line SGC7901, which overexpresses hTERT30 and c-Myc genes31 and harbors wild-type Ras genes and p53 genes,32, 33 was purchased from the cell bank of the Chinese Academy of Sciences. HEK293 cells were obtained from Conservation Genetics CAS Kunming Cell Bank. All cells were cultured in medium supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Israel), 100 units/mL penicillin, and 100 μg/mL streptomycin in humidified conditions at 37°C with 5% CO2.
Recombinant oncolytic adenovirus KGHV300, which carried the anti-p21-Ras scFv gene as well as hTERT and HRE double-controlled promoters and the wild-type Ad5 adenovirus cilia gene, was constructed in our laboratory. KGHV300 specifically replicated in tumor cells but could still infect normal cells, so its Ad5 adenovirus cilia gene was replaced with the Ad35 adenovirus cilia protein F35 gene, which encoded the adenovirus receptor for CIK cells. This modified oncolytic adenovirus was named KGHV400, but it lacked anti-p21-RAS scFv. Then, the recombinant oncolytic adenovirus KGHV500 was constructed by inserting the anti-p21-Ras scFv gene into KGHV400. The similarities and differences among the above oncolytic adenoviruses are detailed in Figure S1. All the recombinant adenoviruses were amplified and extracted from low-generation HEK293 cells at a growth density of 50% −70%. The recombinant adenoviruses purified by cesium chloride double-density gradient centrifugation were activated and concentrated by dialysis bag, and their titers were determined by TCID50.
Preparation and Identification of CIK Cells
The CIK cells were prepared according to a previously published method.27, 33 In brief, peripheral blood mononuclear cells (PBMCs) were prepared from healthy volunteers’ blood and incubated in RPMI 1640 medium containing 10% FBS, 25 mM HEPES, 100 U/mL penicillin, and 100 μg/mL streptomycin. Then, 1,000 U/mL recombinant human IFN-γ (PeproTech, 300-02, USA) was added to the culture on day 0. After 24-hr incubation, 50 ng/mL anti-CD3 monoclonal antibody (mAb) (OKT eBioscience, USA), 100 U/mL recombinant human IL-1α (PeproTech, 200-01A, USA), and 300 U/mL recombinant human IL-2 (PeproTech, 200-02, USA) were added. The cells were maintained at 1 × 106 cells/mL at 37°C and 5% CO2 and changed into fresh complete medium supplemented with 300 U/mL IL-2 every 3 days.
CIK cells were harvested on day 14 when the cells reached 90% confluence. CIK cells were collected by centrifugation and embedded in paraffin, and then the paraffin-embedded CIK cells were cut into 4-μm-thick sections and subjected to H&E staining and immunohistochemistry for detection of the expression of CD3 and CD56. Sections were baked at 60°C for 2 hr, deparaffinized, and heated in citrate buffer (10 mmol/L citric acid [pH 6.0]) in a pressure cooker for 5 min. After antigen retrieval, sections were washed with water and PBS three times for 2 min each, respectively. Then, the sections were exposed to 1.5% H2O2 for 5 min to block endogenous peroxidases, and they were incubated with 10% blocking serum (goat serum) at 37°C for 40 min. Afterward, the sections were incubated with mouse anti-CD3 mAb (ZSGB-Bio, ZA0503, China) and anti-CD56 mAb (MXB, Kit-0028, China) overnight at 4°C, then with a secondary goat anti-mouse immunoglobulin G (IgG) antibody conjugated to horseradish peroxidase (ZSGB-Bio, ZB5305, China) for 40 min at 37°C, and developed with diaminobenzidine (DAB) solution. Counterstaining was performed using hematoxylin, and the slides were sealed with neutral balsam. Anti-CD46 mAb (Abcam, EPR4014, UK) was used to detect the expression of CD46 protein on CIK cells by immunohistochemistry.
Recombinant Oncolytic Adenovirus KGHV500 Binds to CIK Cells
CIK cells were cocultured with KGHV500 or KGHV400 adenoviruses at an MOI of 100 in a 24-well plate with RPMI 1640 medium containing 10% FBS (300 μL/well) for 48 hr. Then, the CIK cells were collected, fixed, dehydrated, and embedded in paraffin. Anti-type 5 adenovirus hexon antibody (Novus, NBP2-11638, USA) was used as the primary antibody for immunohistochemistry to observe whether KGHV500 and KGHV400 adenoviruses could bind to CIK cells.
In Vitro Tumor Inhibition Assays
KGHV500 and KGHV400 Adenovirus-Infected Tumor Cells
The SGC7901 cells were incubated with KGHV500 and KGHV400 adenovirus at an MOI of 100 and collected by centrifugation after fluorescence and CPE were observed. Then, some of the cells were collected, fixed, dehydrated, and paraffin embedded, and anti-CD46 mAb (Abcam, EPR4014, UK) was used as the primary antibody to detect the expression of CD46 protein on SGC7901 cell membranes. The other cells were fixed with 3.5% glutaraldehyde and then 1% osmic acid for 1–2 hr, followed by graded dehydration in 50%, 60%, 70%, 80%, and 90% ethanol; a mixture of ethanol and acetone; and 100% acetone. The samples were impregnated with epoxy resin for 1.5 hr, embedded, sectioned, and stained with uranyl acetate and lead citrate. All samples were sectioned with an EM UC7 Leica ultrathin microtome (Leica, Germany) and viewed for the KGHV500 and KGHV400 adenovirus particles with a JEM-1400 projection electron microscope (Japan Electronics).
MTT Assays
SGC7901 cells were plated at a density of 5 × 104 cells/well in 96-well plates in 100 μL RPMI 1640 medium supplemented with 10% FBS and cultured for 24 hr at 37°C. Then, the cells were infected with KGHV500 or KGHV400 adenovirus at an MOI of 100. PBS was added to the control group. After infection for 1, 2, 3, 4, or 5 days, 20 μL MTT (5 mg/mL, Amresco, M81801, USA) was added to each well and incubated for 4 hr at 37°C, and then the supernatant was discarded. Then, 150 μL DMSO was added to each well, and the 96-well plates were shaken for 10 min. The absorbance of each well was measured at 490 nm using a microplate reader (Bio-Rad, Model 680, USA).
Scratch Test
SGC7901 cells in the logarithmic growth phase were collected and seeded in a 6-well plate (1 × 106 cells per well), and they were incubated at 37°C in RPMI 1640 medium supplemented with 10% FBS until the cells reached 95% confluence. Micro-pipette tips (20 μL) were used to make vertical scratches in the 6-well plate. PBS was used to remove the falling cells, and the remaining cells were cultured in serum-free RPMI 1640 medium. The KGHV500 and KGHV400 adenoviruses were added to the experimental groups, and an equal volume of PBS was added to the PBS group. At 0, 24, and 48 hr after scratching, 3 fields were selected in each group and photographed to compare the scratch-healing differences, which represent cell migration and healing abilities, among the above groups.
Transwell Assays
The Transwell chambers (3422, Corning, USA) were divided into upper and lower layers with an 8-μm aperture polycarbonate microporous membrane. The Matrigel (BD Matrigel Matrix, 354234, USA) was diluted with serum-free RPMI 1640 medium, and then it was added to the upper layer of the Transwell chamber (60 μL/well) and allowed to polymerize at 37°C for 30 min. The cell density was adjusted to 5 × 104 cells/mL, and the cells were incubated in a 24-well plate until the cells reached 80% confluence. The cells were infected with the KGHV500 and KGHV400 adenoviruses at an MOI of 100 with 3 chambers per group, and PBS was added to the control group.
At 24 hr after transfection, the cells were collected and resuspended in a single-cell suspension with RPMI 1640 medium supplemented with 2% FBS. After that, 100 μL single-cell suspension (3 × 104 cells/mL–1 × 105 cells/mL) was added to the upper chamber, 500 μL RPMI 1640 medium containing 20% FBS was added to the lower chamber, and the chambers were kept in a 5% CO2 incubator at 37°C for 48 hr. Afterward, the chambers were removed and rinsed in PBS; the cells in the upper layer of the microporous membrane were removed carefully using cotton buds, and the cells in the lower layer of the chamber were fixed with 95% methanol solution at room temperature for 15 min and stained with hematoxylin solution for 15 min. The Transwell chambers were rinsed with distilled water, dried naturally, and photographed and observed with an inverted optical microscope. Five fields were selected randomly, and the number of transmembrane cells was counted. The experiment was repeated 3 times.
TUNEL Assays
A TUNEL assay was performed to investigate tumor cell apoptosis. The SGC7901 cells were treated with KGHV500, KGHV400, or PBS for 24 hr and then embedded in wax blocks for sectioning. TUNEL assays were performed with sections using an In Situ Cell Death Detection kit, POD (Roche, 11684817910, Switzerland), according to the manufacturer’s instructions. The apoptosis of SGC7901 cells was visualized using a fluorescence microscope, and the percentages of positive cells in five randomly selected high-power fields were calculated.
In Vivo Tumor Inhibition Detection
SGC7901 Tumor Xenograft Model and Tumor Measurement
Specific pathogen-free (SPF) BALB/c-nude mice (female, 4 weeks old) were purchased from Beijing Weitong Lihua Experimental Animal Technology and fed under SPF conditions. Human gastric cancer SGC7901 cells in the logarithmic growth phase were collected, digested with trypsin, centrifuged at 1,000 rpm for 5 min, and resuspended in PBS. SGC7901 cells (1 × 107 cells/200 μL) were subcutaneously inoculated into the right axilla of the BALB/c nude mice. When the average tumor diameter was ≥0.5 cm (usually 2–3 weeks), the mice could be used in this experiment; 40 mice bearing SGC7901 tumor xenografts were assigned randomly to five groups.
Experimental group mice were injected with CIK cells carrying KGHV500 via the tail vein (CIK + KGHV500 group); the other groups were injected with CIK cells carrying KGHV400 (CIK + KGHV400 group), CIK cells, KGHV500, and PBS through the tail vein. The weights of the mice in each group were recorded before and after treatment. After 34 days, the skin on the tumor tissues of the mice in the KGHV500 and PBS groups ulcerated and bled, so all the mice were euthanized and the growth curves were drawn. The tumor diameters (long diameter and short diameter) were measured with a Vernier caliper every 2 days, and the tumor volumes were calculated using the following formula: V = 1/2ab2 (a and b indicated the tumor’s long diameter and short diameter).
Histopathology and Immunohistochemistry
To observe the distribution of KGHV500 adenovirus in nude mice, the mice were sacrificed at 1, 2, 3, 5, or 7 days. Then the mice were dissected. The tumor tissue and all organs (heart, liver, spleen, lungs, kidneys, stomach, pancreas, large intestine, small intestine, and brain) were fixed in formalin, paraffin embedded, sectioned, and stained with H&E. Histological analysis of all the organs was performed by a pathologist using a microscope (Olympus, BX41, Japan). A TUNEL assay was used to detect cell apoptosis in tumor tissue. Immunohistochemistry was employed to detect the expression of adenovirus hexon and anti-p21 RAS scFv in tumors and organs. The anti-FLAG tag antibody (Abnova, MAB9744, China) and anti-human adenovirus hexon mouse mAb (Novus, NBP211638, USA) were used to mark anti-p21 RAS scFv and KGHV500 adenovirus, respectively.
Western Blot Assay
Western blotting was used to detect the expression of anti-p21-Ras scFv in tumors from nude mice. Briefly, total proteins were extracted from fresh tumor tissues in the CIK + KGHV500 group and the KGHV500 group. The total proteins were electrophoresed on SDS-PAGE gels, transferred to polyvinylidene fluoride (PVDF) membranes at 4°C for 2 hr, and then blocked with 5% fat-free milk. The membrane was incubated with the anti-FLAG tag mouse mAb or the mouse anti-β-actin mAb (ZSGB-Bio, TA-09, China), followed by incubation with goat anti-mouse IgG and horseradish peroxidase (HRP) (ZSGB-Bio, ZB-5305, China) and DAB solution. Finally, the expression of the anti-p21-RAS scFv was estimated by the depth and width of the color band.
qPCR Detection of Apoptotic Genes
qPCR was performed to observe the expression of pro-apoptotic genes (caspase3, caspase7, and p53) and anti-apoptosis genes (Bcl-2 and survivin). Briefly, tumor tissue and all the organs were frozen with liquid nitrogen, ground into powder, and rapidly transferred to nuclease-free 1.5-mL Eppendorf (EP) tubes. Then, 100 μL lysis solution (supplemented with β-mercaptoethanol) was added to the EP tubes and allowed to fully mix until there was no obvious tissue bulk in the lysate. Total RNA was extracted according to the instructions of the Eastep Super Total RNA Extraction Kit (Promega, LS1040, USA) with gDNA Remover Master Mix. Then, RNA was reverse transcribed into cDNA using the Eastep RT Master Mix Kit (Promega, LS2050, USA), and the qPCR reaction was performed on an instrument in accordance with the protocol of GoTaq qPCR Master Mix (Promega, A6001, USA). The reaction conditions were as follows: 95°C for 3 min; 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s (45 cycles); 72°C for 7 min; and 4°C for 1 min. The different samples of the treatment group were mixed together, diluted at least 5 gradients, and reacted following the above protocol to establish the standard curve. Finally, the results were analyzed by the standard curve automatically generated by the instrument.
Statistical Analysis
All statistical analyses were performed using SPSS version 19.0. All the measurement data were described as the mean ± SD ( ± s). Comparisons of multiple groups were performed with one-way ANOVA. Statistical significance was indicated by a p value less than 0.05.
Author Contributions
Conceptualization, J.Y. and Y.H.; Investigation, Y.H., S.S., J.C., J.L., and H.F.; Resources, Q.F. and X.P.; Writing – Original Draft, M.W.; Supervision, X.P., Q.F., and J.Y.; Funding Acquisions, J.Y.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81460464) and the Major Science and Technology Project of the Yunnan Science and Technology Plan (2018ZF009).
Footnotes
Supplemental Information includes two figures and one table and can be found with this article online at https://doi.org/10.1016/j.omto.2018.10.003.
Supplemental Information
References
- 1.Ang T.L., Fock K.M. Clinical epidemiology of gastric cancer. Singapore Med. J. 2014;55:621–628. doi: 10.11622/smedj.2014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park S.C., Chun H.J. Chemotherapy for advanced gastric cancer: review and update of current practices. Gut Liver. 2013;7:385–393. doi: 10.5009/gnl.2013.7.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paoletti X., Oba K., Burzykowski T., Michiels S., Ohashi Y., Pignon J.P., Rougier P., Sakamoto J., Sargent D., Sasako M., GASTRIC (Global Advanced/Adjuvant Stomach Tumor Research International Collaboration) Group Benefit of adjuvant chemotherapy for resectable gastric cancer: a meta-analysis. JAMA. 2010;303:1729–1737. doi: 10.1001/jama.2010.534. [DOI] [PubMed] [Google Scholar]
- 4.Bang Y.J., Van Cutsem E., Feyereislova A., Chung H.C., Shen L., Sawaki A., Lordick F., Ohtsu A., Omuro Y., Satoh T., ToGA Trial Investigators Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 5.De Vita F., Giuliani F., Silvestris N., Catalano G., Ciardiello F., Orditura M. Human epidermal growth factor receptor 2 (HER2) in gastric cancer: a new therapeutic target. Cancer Treat. Rev. 2010;36(Suppl 3):S11–S15. doi: 10.1016/S0305-7372(10)70014-1. [DOI] [PubMed] [Google Scholar]
- 6.Milburn M.V., Tong L., deVos A.M., Brünger A., Yamaizumi Z., Nishimura S., Kim S.H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 7.Young A., Lyons J., Miller A.L., Phan V.T., Alarcón I.R., McCormick F. Ras signaling and therapies. Adv. Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 8.Papke B., Der C.J. Drugging RAS: Know the enemy. Science. 2017;355:1158–1163. doi: 10.1126/science.aam7622. [DOI] [PubMed] [Google Scholar]
- 9.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J.L., Liu D.X., Zhen S.J., Zhou Y.G., Zhang D.J., Yang L.Y., Chen H.B., Feng Q. A novel anti-p21Ras scFv antibody reacting specifically with human tumour cell lines and primary tumour tissues. BMC Cancer. 2016;16:131. doi: 10.1186/s12885-016-2168-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J.L., Pan X.Y., Zhao W.X., Hu Q.C., Ding F., Feng Q., Li G.Y., Luo Y. The antitumor efficacy of a novel adenovirus-mediated anti-p21Ras single chain fragment variable antibody on human cancers in vitro and in vivo. Int. J. Oncol. 2016;48:1218–1228. doi: 10.3892/ijo.2016.3334. [DOI] [PubMed] [Google Scholar]
- 12.Pan X.Y., Liu X.J., Li J., Zhen S.J., Liu D.X., Feng Q., Zhao W.X., Luo Y., Zhang Y.L., Li H.W., Yang J.L. The antitumor efficacy of anti-p21Ras scFv mediated by the dual-promoter-regulated recombinant adenovirus KGHV300. Gene Ther. 2017;24:40–48. doi: 10.1038/gt.2016.74. [DOI] [PubMed] [Google Scholar]
- 13.Introna M. CIK as therapeutic agents against tumors. J. Autoimmun. 2017;85:32–44. doi: 10.1016/j.jaut.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Gao X., Mi Y., Guo N., Xu H., Xu L., Gou X., Jin W. Cytokine-Induced Killer Cells As Pharmacological Tools for Cancer Immunotherapy. Front. Immunol. 2017;8:774. doi: 10.3389/fimmu.2017.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng N., Zhao X. Comparison of K-ras mutations in lung, colorectal and gastric cancer. Oncol. Lett. 2014;8:561–565. doi: 10.3892/ol.2014.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasper H.-U., Schneider-Stock R., Mellin W., Roessner A. P21 protein expression and ras-oncogene mutations in gastric carcinoma: correlation with clinical data. Int. J. Oncol. 1998;12:69–74. doi: 10.3892/ijo.12.1.69. [DOI] [PubMed] [Google Scholar]
- 17.Fujita K., Ohuchi N., Yao T., Okumura M., Fukushima Y., Kanakura Y., Kitamura Y., Fujita J. Frequent overexpression, but not activation by point mutation, of ras genes in primary human gastric cancers. Gastroenterology. 1987;93:1339–1345. doi: 10.1016/0016-5085(87)90264-2. [DOI] [PubMed] [Google Scholar]
- 18.Bai S., Feng Q., Pan X.Y., Zou H., Chen H.B., Wang P., Zhou X.L., Hong Y.L., Song S.L., Yang J.L. Overexpression of wild-type p21Ras plays a prominent role in colorectal cancer. Int. J. Mol. Med. 2017;39:861–868. doi: 10.3892/ijmm.2017.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong Y.L., Yang L.Y., Pan X.Y., Feng Q., Zou H., Song S.L., Wang L., Wang P., Bai S., Zhou X.L., Yang J.L. Mutation status of ras genes in breast cancers with overexpressed p21Ras protein. Int. J. Clin. Exp. Pathol. 2016;9:10422–10429. [Google Scholar]
- 20.Shi M., Shi H., Ji J., Cai Q., Chen X., Yu Y., Liu B., Zhu Z., Zhang J. Cetuximab inhibits gastric cancer growth in vivo, independent of KRAS status. Curr. Cancer Drug Targets. 2014;14:217–224. doi: 10.2174/1570163811666140127145031. [DOI] [PubMed] [Google Scholar]
- 21.Chira S., Jackson C.S., Oprea I., Ozturk F., Pepper M.S., Diaconu I., Braicu C., Raduly L.Z., Calin G.A., Berindan-Neagoe I. Progresses towards safe and efficient gene therapy vectors. Oncotarget. 2015;6:30675–30703. doi: 10.18632/oncotarget.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W.W., Li L., Li D., Liu J., Li X., Li W., Xu X., Zhang M.J., Chandler L.A., Lin H. The First Approved Gene Therapy Product for Cancer Ad-p53 (Gendicine): 12 Years in the Clinic. Hum. Gene Ther. 2018;29:160–179. doi: 10.1089/hum.2017.218. [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Li B., Li C.J., Li L.J. Key points of basic theories and clinical practice in rAd-p53 ( Gendicine ™) gene therapy for solid malignant tumors. Expert Opin. Biol. Ther. 2015;15:437–454. doi: 10.1517/14712598.2015.990882. [DOI] [PubMed] [Google Scholar]
- 24.Fukuhara H., Ino Y., Todo T. Oncolytic virus therapy: A new era of cancer treatment at dawn. Cancer Sci. 2016;107:1373–1379. doi: 10.1111/cas.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufman H.L., Kim D.W., DeRaffele G., Mitcham J., Coffin R.S., Kim-Schulze S. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann. Surg. Oncol. 2010;17:718–730. doi: 10.1245/s10434-009-0809-6. [DOI] [PubMed] [Google Scholar]
- 26.Howells A., Marelli G., Lemoine N.R., Wang Y. Oncolytic Viruses-Interaction of Virus and Tumor Cells in the Battle to Eliminate Cancer. Front. Oncol. 2017;7:195. doi: 10.3389/fonc.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt-Wolf I.G., Negrin R.S., Kiem H.P., Blume K.G., Weissman I.L. Use of a SCID mouse/human lymphoma model to evaluate cytokine-induced killer cells with potent antitumor cell activity. J. Exp. Med. 1991;174:139–149. doi: 10.1084/jem.174.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mata-Molanes J.J., Sureda González M., Valenzuela Jiménez B., Martínez Navarro E.M., Brugarolas Masllorens A. Cancer Immunotherapy with Cytokine-Induced Killer Cells. Target. Oncol. 2017;12:289–299. doi: 10.1007/s11523-017-0489-2. [DOI] [PubMed] [Google Scholar]
- 29.Thorne S.H., Negrin R.S., Contag C.H. Synergistic antitumor effects of immune cell-viral biotherapy. Science. 2006;311:1780–1784. doi: 10.1126/science.1121411. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y.B., Guo L.P., Yao P., Ning X.Y., Aerken G., Fang D.C. Telomerase and hTERT: can they serve as markers for gastric cancer diagnosis? World J. Gastroenterol. 2014;20:6615–6619. doi: 10.3748/wjg.v20.i21.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Hou Y., Ashktorab H., Gao L., Xu Y., Wu K., Zhai J., Zhang L. The impact of C-MYC gene expression on gastric cancer cell. Mol. Cell. Biochem. 2010;344:125–135. doi: 10.1007/s11010-010-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji W., Ma J., Zhang H., Zhong H., Li L., Ding N., Jiao J., Gao Z. Role of p53β in the inhibition of proliferation of gastric cancer cells expressing wild-type or mutated p53. Mol. Med. Rep. 2015;12:691–695. doi: 10.3892/mmr.2015.3370. [DOI] [PubMed] [Google Scholar]
- 33.Finke S., Trojaneck B., Lefterova P., Csipai M., Wagner E., Kircheis R., Neubauer A., Huhn D., Wittig B., Schmidt-Wolf I.G. Increase of proliferation rate and enhancement of antitumor cytotoxicity of expanded human CD3+ CD56+ immunologic effector cells by receptor-mediated transfection with the interleukin-7 gene. Gene Ther. 1998;5:31–39. doi: 10.1038/sj.gt.3300560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.