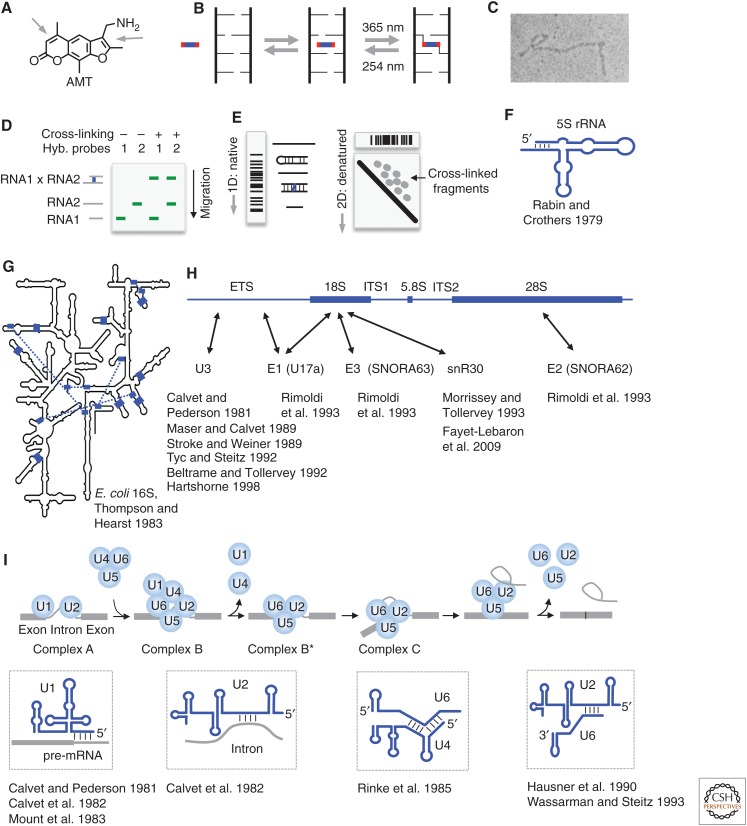
Figure 1.
Psoralen chemistry and historical applications. (A) Structure of the commonly used psoralen AMT (4′-aminomethyltrioxsalen). Arrows point to the double bonds that undergo cycloaddition to pyrimidines. (B) Model of psoralen intercalation into nucleic acid helices and reversible cross-linking. (C) Detection of psoralen cross-linked RNA structures by electron microscopy. (Image courtesy of Paul L. Wollenzien et al.; reprinted, with permission, from Wollenzien et al. 1978, © National Academy of Sciences.) (D) Detection of RNA–RNA interactions using gel electrophoresis and northern blotting. Cross-linked RNA pairs can be identified by the simultaneous hybridization of probes to both RNAs. (E) The “native-denatured” 2D gel method for selecting psoralen cross-linked RNA. The first-dimension gel is native. In the second dimension, the first-dimension gel slices are embedded in a urea-denatured gel and run at high temperature, in which the cross-linked fragments assume an “X” shape and migrate much more slowly, above the diagonal, forming scattered dots for simple RNA species or a smear for complex RNA mixtures, such as total RNA. (Adapted from Lu et al. 2016, with permission, from Elsevier.) (F) Secondary structure of the Escherichia coli 5S ribosomal RNA (rRNA), based on psoralen cross-linking and enzymatic sequencing (Rabin and Crothers 1979). (G) Psoralen cross-linked duplexes (blue blocks and the blue dashed lines) superimposed on the Escherichia coli 16S rRNA model derived from crystal structure (Petrov et al. 2014). (Redrawn, with permission, from Thompson and Hearst 1983.) (H) Summary of small nucleolar RNA (snoRNA)–rRNA interactions based on psoralen cross-linking. ETS/ITS: external/internal transcribed sequence. Arrows point to cross-linking sites. (I) Summary of several RNA–RNA interactions in precursor messenger RNA (pre-mRNA) splicing based on psoralen cross-linking. (The splicing model was redrawn, with permission, from Will and Luhrmann 2011.)