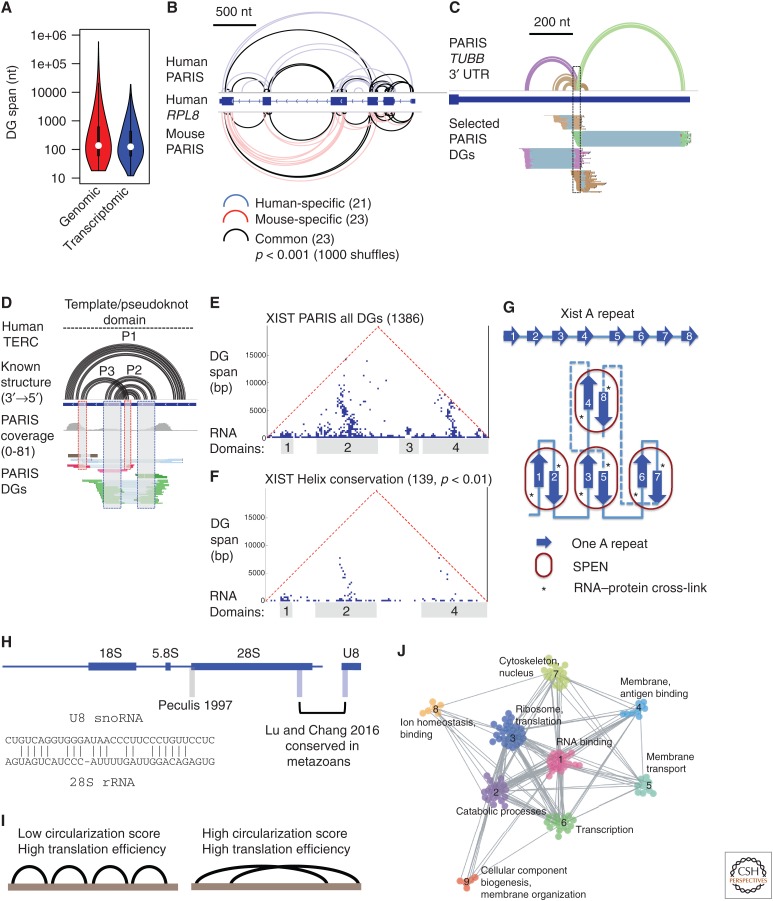
Figure 4.
New insights into the RNA structurome and interactome. (A) Long-range structures are prevalent in the transcriptome. Duplex group (DG) spans were calculated from Henrietta Lacks cell line psoralen analysis of RNA interactions and structures (HeLa PARIS) data on genomic and transcriptomic coordinates. (B) Example of conserved structures in RPL8 messenger RNA (mRNA) based on direct comparison of human (human embryonic kidney cells 293 [HEK293]) and mouse (J1 embryonic stem cells) PARIS data. Significance of the overlap was tested using random shuffling of DGs in the exons. (C) Example of alternative/dynamic structures in the 3′ untranslated region (UTR) of tubulin β class 1 (TUBB) mRNA from HeLa PARIS data. The corresponding structure models (first track) and DGs are color coded. (D) PARIS detects pseudoknot structure (interlocked DGs) in the telomerase RNA component (TERC). (E) PARIS determines the architecture of X-inactive specific transcript (XIST). Each point in the heat map shows the connection between the two regions indicated by the feet of the triangle. Four major domains are obvious from the clustered DGs. (F) About 10% of PARIS-determined duplexes are conserved in eutherian mammals (p value < 0.01). (G) Model of SPEN-A-repeat complex in the XIST ribonucleoprotein (RNP). The base-pairing interactions among the 8.5 repeats are stochastic and only one specific family of conformations from PARIS data is shown here. SPEN binding involves both single- and double-stranded regions but is only cross-linked to the single-stranded region 3–5 nt upstream of the interrepeat duplex unit. (H) PARIS in human and mouse cells revealed the precise U8:28S interaction, which is conserved in metazoans. PARIS-determined interaction sites in blue, and the previously reported site in gray (Peculis 1997). The base-pairing model is shown in the inset. (I) Long-span structures as detected by sequencing of psoralen cross-linked, ligated, and selected hybrids (SPLASH) correlates with higher translation efficiency. (J) RNAs of related functions tend to interact more frequently than nonrelated ones. Global RNA–RNA interaction networks can be organized into modules and the network topology changes in response to stem cell differentiation. (Panels A–H are adapted from Lu et al. 2016; panels I–J are adapted from Aw et al. 2016, both, with permission, from Elsevier.)