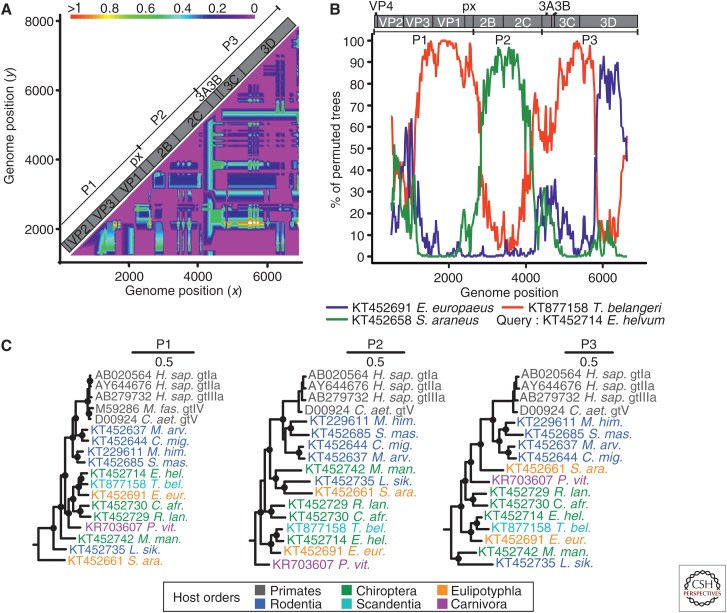
Figure 3.
Evidence of recombination in hepatoviruses. (A) Phylogenetic compatibility scan of the full polyprotein genes of hepatoviruses isolated from humans, rodents, tree shrew, and bats. GenBank Accession numbers of sequences used were: AB279735; KT452644; KT452685; KT452729; KT877158; KT452742; KT452735. The graph was created using SSE 1.3 (Simmonds 2012), a sliding window of 500 nt, and a step size of 50 nt, with a bootstrap cutoff of 70%. (B) The bootscan graph shows percent of bootstrap replicates (y axis) that support grouping of the query sequence with each of three test sequences in a 1000 nt window sliding over the genome at a 20 nt step (x axis, window center position); the plot was done using Simplot 3.5 (Lole et al. 1999) and the Kimura substitution model; dotted line shows the 70% reliable bootstrap support cutoff. As a result of alignment shifts, the genome plots do not precisely correspond to raw genome positions. (C) Bayesian phylogenies of hepatovirus domains P1, P2 (only 2C), and P3 (only 3CD) showing reliable evidence of several recombination events. Viruses are colored according to their host order. Phylogenies were generated at the amino acid level from translation alignments excluding all ambiguous data or gaps using MrBayes V3.1 (Huelsenbeck and Ronquist 2001) and a WAG amino acid substitution model. Trees were generated as described above and rooted by the avian encephalomyelitis virus (genus Tremovirus). Bayesian posterior probabilities above 0.9 are marked by filled circles at nodes. The scale bar indicates genetic distance. M. fas., Macaca fascicularis; C. aet., Chlorocebus aethiops; M. arv., Microtus arvalis; C. mig., Cricetulus migratorius; M. him., Marmota himalayana; S. mas., Sigmodon mascotensis; E. hel., Eidolon helvum; T. bel., Tupaia belangeri chinensis; E. eur., Erinaceus europeaensis; C. afr., Coleura afra; R. lan., Rhinolophus landeri; P. vit., Phoca vitulina; M. man., Miniopterus cf. manavi; L. sik., Lophuromys sikapusi; S. ara., Sorex araneus.