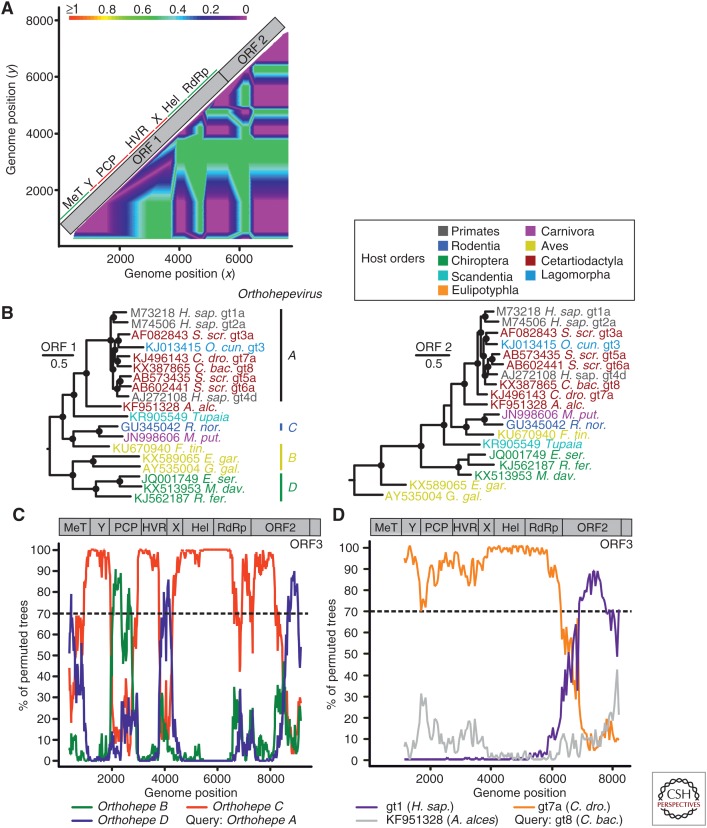
Figure 4.
Evidence of recombination in hepeviruses. (A) Phylogenetic compatibility scan of concatenated open reading frame (ORF)1 + ORF2 alignment of hepeviruses isolated from human (X98292), tree shrew (KR905549), Falco tinnunculus (KU670940), Rattus rattus (AB847306), chicken (KF511397), Eptesicus serotonius (JQ001749), and Rhinolophus ferrumequinum (KJ562187) was generated as described in the legend to Figure 3A. (B) Bayesian phylogenies of complete ORF1 and 2 of orthohepeviruses. Viruses are colored according to their host order. Phylogenies were generated at the nucleotide level from translation alignments excluding all ambiguous data or gaps using MrBayes V3.1. A general time-reversible (GTR) model with a γ distribution (G) across sites and a proportion of invariant sites (I) (GTR + G + I) was used as the substitution model. Trees were generated as above. Trees were rooted by the sister genus Piscihepevirus. Bayesian posterior probabilities above 0.9 are marked by filled circles at nodes. The scale bar indicates genetic distance. C. bac., Camelus bactrianus; A. alc., Alces alces; R. nor., Rattus norvegicus; M. put., Mustela putorius furo; F. tin., Falco tinnunculus; E. gar., Egretta garzetta; E. ser., Eptesicus serotinus; G. gal., Gallus gallus; M. dav., Myotis davidii; R. fer., Rhinolophus ferrumequinum. (C) Evidence of recombination in the species Orthohepevirus A relative to species B–D. The scan was performed similarly as described in the legend for Figure 3B. Window size was 800 nt, step size at 20 nt. Accession numbers of sequences used: JQ013793; JN998606; AY535004; KJ562187. As a result of alignment shifts, the genome plots do not precisely correspond to raw genome positions. Orthohepe, Orthohepevirus. (D) Evidence of recombination in species Orthohepevirus A genotype (gt) 8 (C. bactrianus) relative to gt1 (Homo sapiens), gt7a (Camelus dromedarius), and the moose (A. alces) hepevirus. Window size was 2500 nt, step size 50 nt. Accession numbers of sequences used: KX387865; KJ496143; X98292; KF951328. ORF1–ORF3 were concatenated for bootscan analyses, as indicated by genomic representations above panels C and D. Hel, Helicase; HVR, hypervariable genome region; PCP, papain-like cysteine protease.