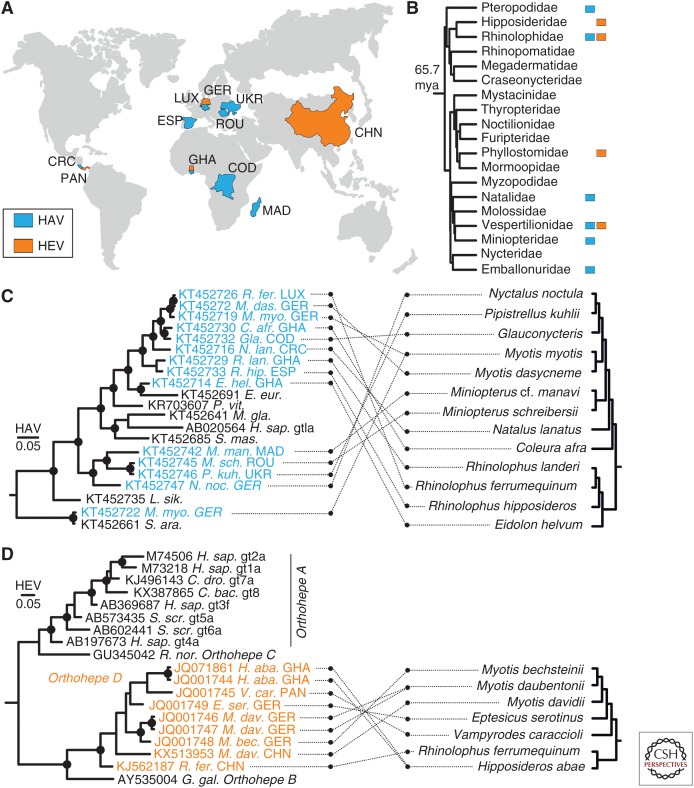
Figure 5.
Global distribution and phylogenetic relationships of bat hepatoviruses and hepeviruses. (A) Map showing the geographic origins of recently identified bat hepatoviruses (blue) and bat hepeviruses (orange). CHN, China; COD, Democratic Republic of the Congo; CRC, Costa Rica; ESP, Spain; GER, Germany; GHA, Ghana; LUX, Luxembourg; MAD, Madagascar; PAN, Panama; ROU, Romania; UKR, Ukraine. (B) Chiropteran phylogeny (phylogeny adapted from Foley et al. 2016) complemented manually by the family of Miniopteridae, which diverged around 43 million years ago (mya) from the Vespertilionidae (Miller-Butterworth et al. 2007). Bat families in which hepatitis A virus (HAV)- or hepatitis E virus (HEV)-related viruses have been found are tagged with blue or orange squares, respectively. (C) Bayesian phylogeny of a 863-nucleotide partial VP2/VP3 region of bat and representative nonchiropteran hepatoviruses. Analyzed region corresponds to positions 391–1253 in a prototype genotype (gt) Ia HAV strain (GenBank AB020564). M. gla., Myodes glareolus. See also the legend to Figure 3. (D) Bayesian phylogeny of a 324-nucleotide partial RNA-dependent RNA polymerase (RdRp) region of bat and representative nonchiropteran hepeviruses. The analyzed region corresponds to positions 4255–4577 in an HEV gt1 prototype strain (GenBank accession number M73218). Generally, Bayesian phylogenies were generated on translation alignments excluding all ambiguous data or gaps using MrBayes V3.1 (Huelsenbeck and Ronquist 2001) and a WAG amino acid substitution matrix. Trees were rooted by Tremovirus for HAV and Piscihepevirus for HEV, respectively. Bayesian posterior probabilities above 0.9 are marked by filled circles at nodes. The scale bar indicates genetic distance. Host trees were generated using complete cytochrome B coding sequences retrieved from GenBank and settings as described for virus phylogenies, including priors, to increase phylogenetic resolution above the level of host families.