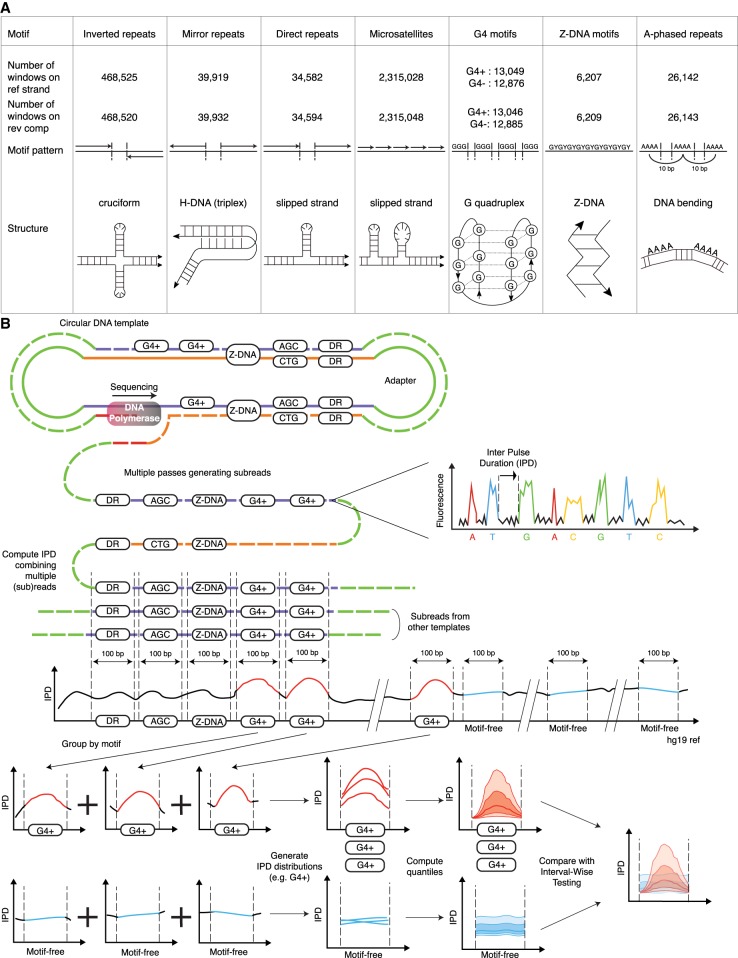
Figure 1.
Non-B DNA motifs and Inter-Pulse Duration (IPD) analysis pipeline. (A) Non-B DNA motif types: patterns, putative structures, and counts of nonoverlapping 100-bp windows containing one (and only one) motif with IPD measurements on the reference or reverse-complement strand. (B) During SMRT sequencing, IPDs are recorded for each nucleotide in each subread (each pass on the circular DNA template). Subreads are aligned against the genome, and an IPD value is computed averaging ≥3 subread IPDs for each genome coordinate. We form 100-bp windows around annotated non-B motifs, extract their IPD values, and pool windows containing motifs of the same type to produce a distribution of IPD curves over the motif and its flanks (represented via 5th, 25th, 50th, 75th, and 95th quantiles along the 100 window positions). We also form a set of nonoverlapping 100-bp windows free from any known non-B motif and pool them to produce a “motif-free” distribution of IPD curves. Each motif type is then compared to motif-free windows through Interval-Wise Testing (IWT). (G4) G-quadruplexes; (G4+/G4−) G4 annotated on the reference/reverse-complement strand; (DR) direct repeats.