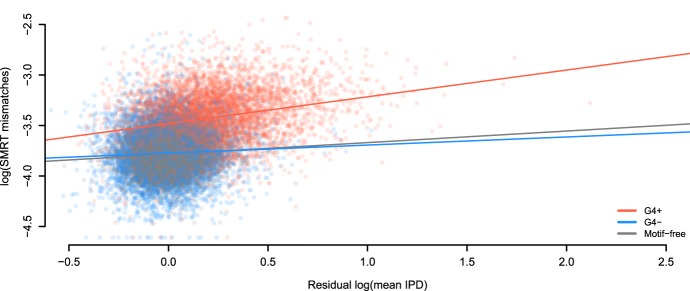
Figure 4.
Errors are linked to kinetic variation. Sequencing through templates containing G-quadruplex motifs (G4+) increases the positive relationship between mismatches in SMRT sequencing and IPD values (corrected for sequence composition). This does not occur when sequencing through the reverse complement of G-quadruplex motifs (G4−). The regression model accounting for three groups of regions (G4+, G4−, and motif-free windows) has R2 equal to 35.4%. Slopes are 0.26, 0.08, and 0.11 for G4+ motifs (N = 5937), G4− motifs (N = 5695), and motif-free windows (N = 11,632, which is the sum of samples sizes for G4+ and G4−), respectively.