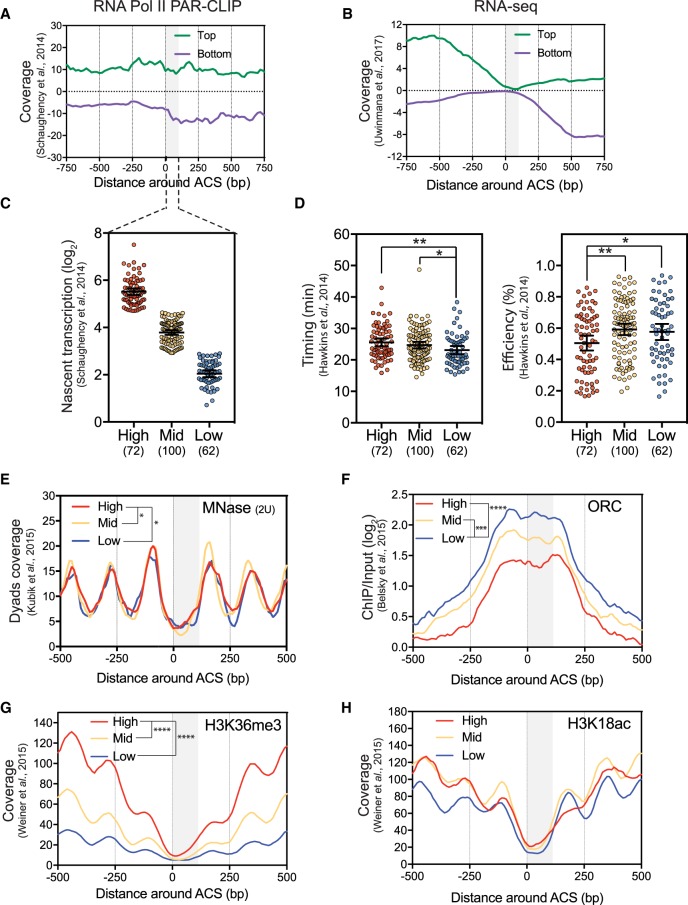
Figure 3.
Natural pervasive transcription correlates with timing/efficiency and specific chromatin features of replication origins. (A) Metagene analysis of Top and Bottom RNA Pol II PAR-CLIP (Schaughency et al. 2014) and (B) RNA-seq (Uwimana et al. 2017) data in the vicinity of 234 oriented replication origins. Data were smoothed by a 20-bp moving window. The gray box represents the window in which transcriptional readthrough was analyzed. (C) The 234 replication origins were divided into three classes (High, Mid, or Low) according to their level of total natural readthrough transcription using a nonbiased k-means clustering approach. This basal nascent transcription was calculated on oriented ARSs by adding the RNA Pol II PAR-CLIP mean densities between the ACS and +100 bp on the top strand to the signal over the same region on the bottom strand. Each 100-bp segment was considered as 1 bin. Natural nascent transcription data were taken from Schaughency et al. (2014). (D) Replication timing and efficiency of the three classes of ARSs defined as in C. (E–H) Nucleosome positioning, ORC, H3K36me3, and H3K18ac levels considering the three classes of replication origins. Data for nucleosome positioning, ORC recruitment, and histone marks were retrieved from Kubik et al. (2015), Belsky et al. (2015), and Weiner et al. (2015), respectively. For these plots, ARSs were oriented and aligned according to their ACS T-rich sequence. The significance of differences between the three classes was calculated over the ACS to +100 bp region considered as 1 bin and is indicated in the top left of each panel. No annotation is considered as nonsignificant. (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001; (****) P-value < 0.0001.