Fig. 1.
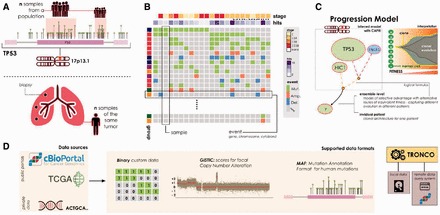
(A) TRONCO can process either alterations (e.g. somatic mutations or wider chromosomal lesions) in a cohort of independent samples (top lolliplot diagram), or a set of multiple snapshots from a unique patient (e.g. multi-region or single-cell, bottom panel). (B) Oncoprints allow the user to visualize the data that the tool is processing. Regardless of the source, each row represents a certain alteration—at a custom resolution depending on the cancer under study—and each column a sample. (C) A model inferred with the tool might outline cancer evolution occurring in a population ensemble or in an individual patient. Graphically, alterations are represented as nodes with different colors (e.g. green mutations and blue homozygous deletions). Algorithms such as CAPRI allow describing alterations with logical formulas, in an attempt to find their role as a ‘group’ (see Ramazzotti et al., 2015 for details); we picture such groups with dashed lines. In the panel, we show a hypothetical ensemble-level model predicting a selection pressure on two genes mapped to 17p13, tp53 and hic1, as it may be inferred by analyzing samples harboring either tp53/hic1 mutations or homozygous deletions in the cytoband where any of these two genes map, i.e. here for purely explanatory cases we suppose just tp53, which maps to 17p13.1. The model suggests a trend of selection toward mutations in gene y, which shall be interpreted as a set of preferential clonal expansions characteristic of the population of analyzed samples, involving alterations of the functions mapped to 17p13 and y. (D) TRONCO supports three data types. Custom data, which is supposed to be provided as a binary input matrix storing the presence (1) or absence (0) of a certain alteration in a sample. Or, standard data formats such as the MAF for somatic mutations, as well as the GISTIC format for focal Copy Number Variations. Data can be generated by custom experiments, or collected—along with other ‘omics’—from public databases such as TCGA and cBio portal. For the latter, cBio portal, TRONCO implements a query system to fetch data with minimal effort. The tool engine can then be used to manipulate genomic profiles—regardless of their source—and run progression inference algorithms
