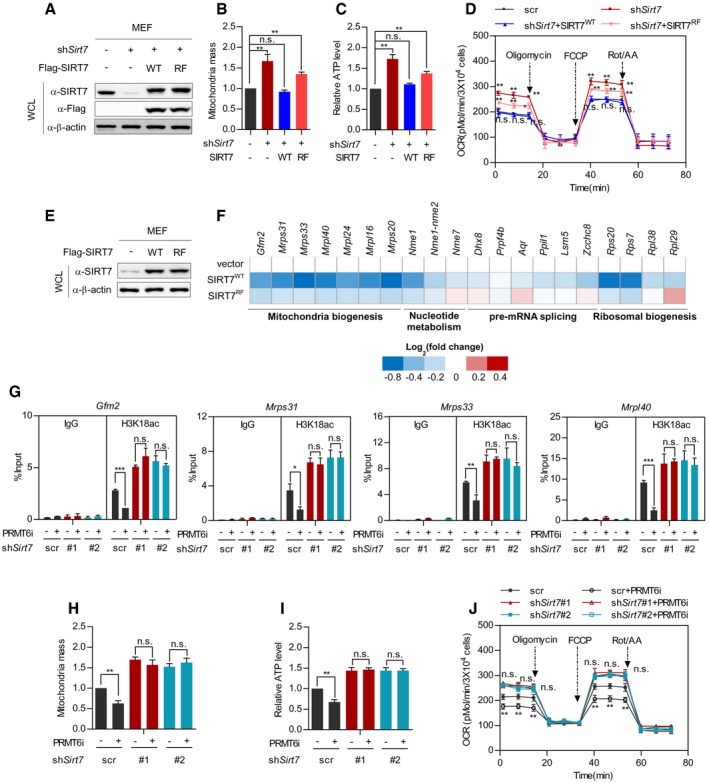
-
A–D
Wild‐type SIRT7, but not its RF mutant, restores mitochondria mass, ATP level, and cellular respiration in Sirt7‐deficient MEF cells. Wild‐type SIRT7 or its RF mutant was re‐expressed in Sirt7‐knockdown MEF cells (A). Mitochondria mass was determined by MitoTracker Red staining (B). Cellular ATP was quantified and normalized to cell number (C). Respiration flux was determined as indicated (D). (mean ± SD, n = 3 experimental replicates, *P < 0.05; **P < 0.01; n.s. = not significant, unpaired two‐tailed t‐test).
-
E, F
Wild‐type SIRT7, but not its RF mutant, downregulates SIRT7‐target genes involved in mitochondria biogenesis. Wild‐type SIRT7 and its RF mutant were stably expressed in MEF cells (E). SIRT7‐target gene expression was determined and presented as a heatmap on a log2 scale (F).
-
G–J
Inhibition of Prmt6 decreases mitochondria mass, ATP level, and the respiration flux of control cells, but not Sirt7‐knockdown MEF cells. Control cells and stable Sirt7‐knockdown MEF cells were treated with or without 5 μM PRMT6i for 24 h. The H3K18ac level at the promoter of SIRT7‐target genes (Gfm2, Mrps31, Mrps33, and Mrpl40) was determined by ChIP‐qPCR, and Rabbit IgG was used as a negative control (G). Mitochondria mass was determined by MitoTracker Red staining (H). Cellular ATP was quantified and normalized to cell number (I). Respiration flux was determined as indicated (J). (Data shown are mean ± SD, n = 3 experimental replicates, *P < 0.05; **P < 0.01; ***P < 0.001; n.s. = not significant, unpaired two‐tailed t‐test)