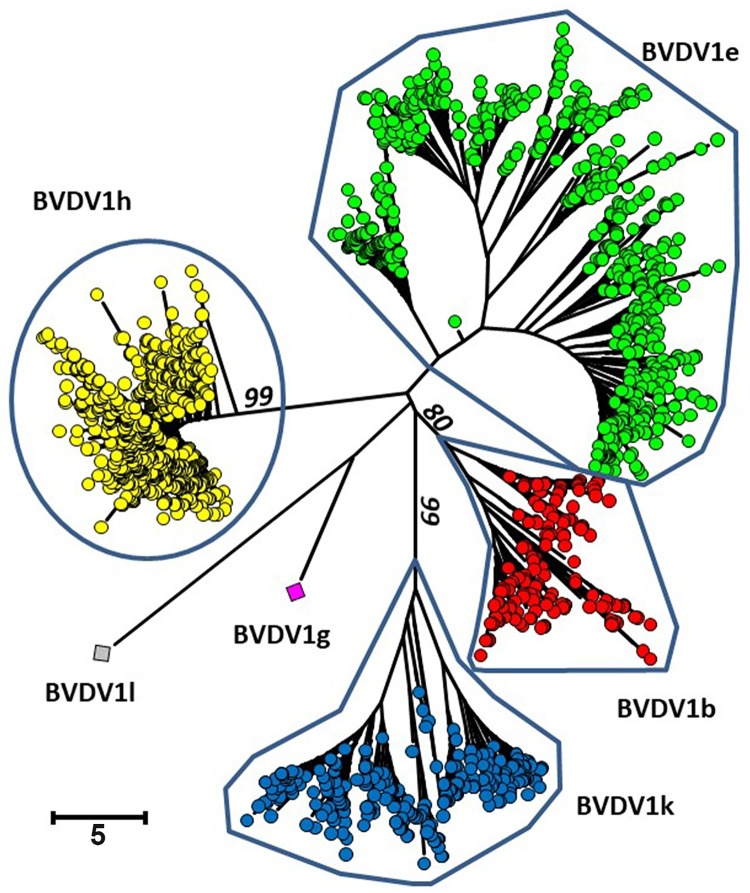
Fig 1. Phylogenetic analysis based on the 5’UTR of the BVD viral genome.
The alignment was performed with the program MUSCLE [38] and the phylogenetic analysis with the program MEGA6 [21]. Duplicate sequences were eliminated. The evolutionary history was inferred using the Neigbor-Joining method [39]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches (only values higher than 80 are shown) [40]. All positions containing gaps and missing data were eliminated. The classification was adapted from [18].