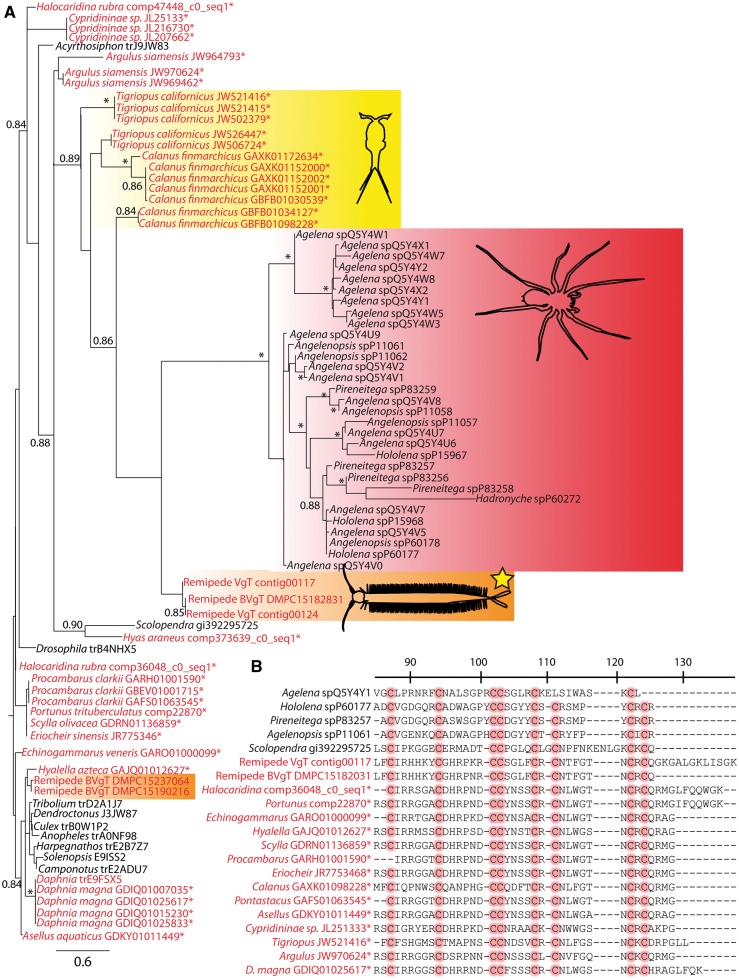
Fig. 5.
(A) Phylogenetic hypothesis of the evolution of the β/δ agatoxin-like domain from putative neurotoxins in crustaceans, based on the approximate ML methods implemented in FastTreeMP (a qualitatively similar tree was generated using RAxML —see Supplementary File S4). Crustacean taxa names are in red; other sequences are in black; new crustacean sequences from the TSA database of NCBI or from unpublished transcriptomic assemblies are indicated with an asterisk (e.g., Hyas and Portunus sequences are from gill tissue). The clade forming known nuerotoxins from spiders is outlined in red, the described neurotoxin from the remipede venom gland is outlined in orange and highlighted with a yellow star, while the “nontoxin” paralog expressed in other remipede tissues is outlined in orange only (no star). The copepod sequences closely related to known neurotoxins are outlined in yellow. Scale bar indicates amino acid replacements per site. Numbers at nodes indicate “SH-like local support values” based on 1000 resamples (Price et al. 2010), with those < 0.8 removed and those > 0.9 indicated with an asterisk. The alignment used in generating this phylogeny and the resulting tree in Newick format are available as Supplementary Files S2 and S3. (B) Representative alignment of the β/δ agatoxin-like domain from putative neurotoxin-like proteins in crustaceans, with functionally important and conserved cysteines highlighted in red. Numbering and other details follow von Reumont et al. (2014).