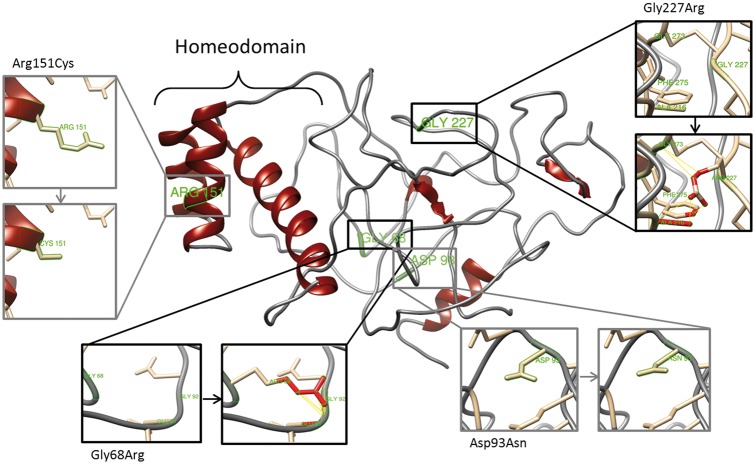
Figure 5.
Effect of non-synonymous mutations on RHOXF2/2B protein structure. Diagram of the top predicted tertiary structure model of RHOXF2/2B using Phyre2. The homeodomain region was modeled with template structures of homeodomain containing proteins and the remaining residues ab initio. The model was colored by secondary structure using UCSF Chimera software: helix structures are shown in red and coiled structures are shown in grey. Each insert shows the identified non-synonymous mutations (grey) and SNPs (black) as sticks. Predicted clashes/contacts for c.202G > A (p.Gly68Arg) and c.679G > A (p.Gly227Arg) are marked with yellow lines (14 and 5, respectively) and atoms involved in the potential contacts are highlighted in red (10 and 8, respectively). No clashes/contacts were predicted for c.277G > A (p.Asp93Asn) and c.451C > T (p.Arg151Cys).