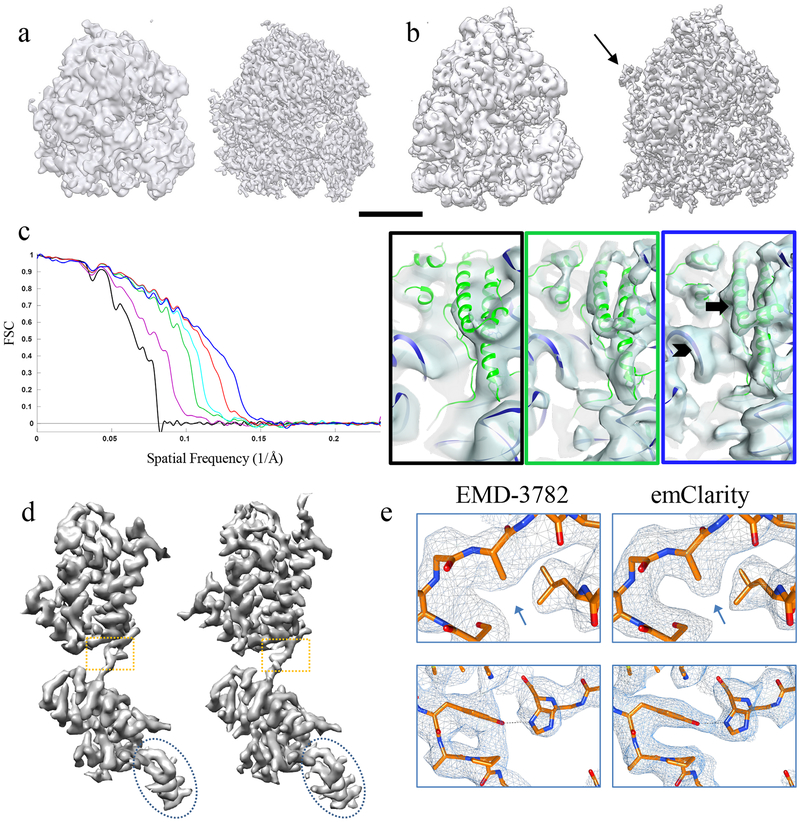
Figure 2. Improvement in resolution of sub-tomogram averaging using emClarity.
(a-b) Comparison of the sub-tomogram average of yeast 80s ribosome (a) and of rabbit 80s ribosome (b) by RELION (EMD-3228) (a, left) or by pyTOM (EMD-3420) (b, left) and by emClarity (right). Arrow points to an additional feature only revealed in emClarity. Scale bar, 100 Å. (c) Cross-FSC between the sub-tomogram averages by emClarity and the SPA cryoEM map (EMD-2275) of yeast 80s ribosome, each accumulating the previous improvement: original orientation parameters from Relion (black), CTF estimation and correction with the optimal exposure filter (magenta), one round of tomo-CPR (green), per-tilt defocus estimation (cyan), consideration of resolution anisotropy (red), and with all features plus alignment in emClarity (blue). Right, representative views of sub-tomogram averages, with the frame color matching the plot colors, and a rigid body docking of yeast 80s atomic model (PDB-47VR). The arrow and chevron highlight the resolved alpha helices and RNA structures, respectively. These experiments were repeated at least three times with nominally identical results. (d) Comparison of the sub-tomogram average of HIV-1 immature CA-SP1 monomer (EMD-3782) (left) and by emClarity (right). (e) Enlarged views of boxed area in d overlaid with a real-space refined model.