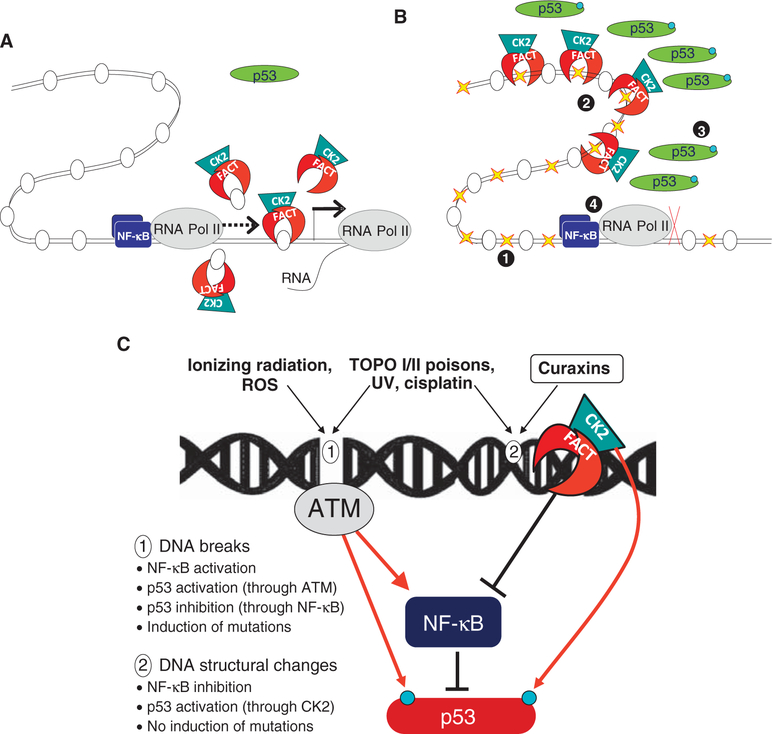
Fig. 8.
Proposed model of curaxins’ mechanism of activity (see details in the text). (A and B) FACT involved in transcription elongation on normal conditions (A) is trapped in chromatin in curaxin-treated cells (B). 1, curaxin binds DNA and changes chromatin architecture; 2, FACT is trapped in chromatin; 3, p53 is phosphorylated by CK2; 4, NF-κB transcription is blocked. (C) Two types of consequences of small-molecule DNA interactions. 1, DNA breaks result from ionizing radiation and reactive oxygen species (ROS) reaction with DNA; 2, Changes in DNA 3D structure caused by nonreactive intercalators such as curaxins. Inhibitors of topoisomerases or compounds causing covalent modifications of DNA may cause structural changes in DNA and breaks. Red arrow, effects leading to an increase in the activity or the targeted factor; black lines, effect leading to an inhibition of the activity of the targeted factor.