Figure 1.
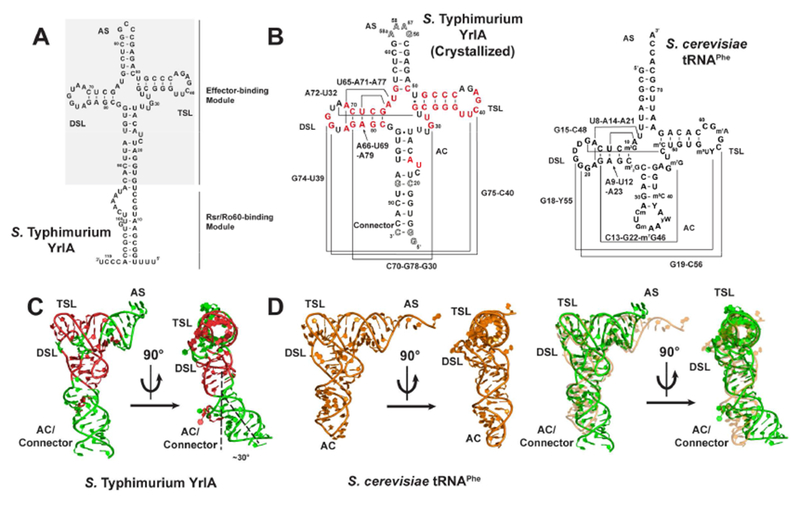
S. Typhimurium YrlA folds into a structure very similar to that of canonical tRNA. (A) Full-length S. Typhimurium YrlA consists of an Rsr/Ro60-binding module and an effector-binding module (shaded) that resembles tRNA. (B) Secondary structures of the crystallized S. Typhimurium YrlA effector-binding module (nt 16-92) and S. cerevisiae tRNAPhe. Important tertiary interactions are connected by lines and are labeled. Invariant or nearly invariant (>80% conserved) nucleotides in YrlA are in red (Chen et al., 2014). Modified nucleotide sequences to facilitate crystallization are shown as hollow characters. (C) The crystal structure of S. Typhimurium YrlA (green/red, PDB: 6cu1). The conserved nucleotides shown in panel B are in red. (D) Left, the tertiary structure of S. cerevisiae tRNAPhe (orange, PDB: 4tna); right, overlay of the YrlA (green) and tRNAPhe (orange) structures. See also Figure S1 and S2.
