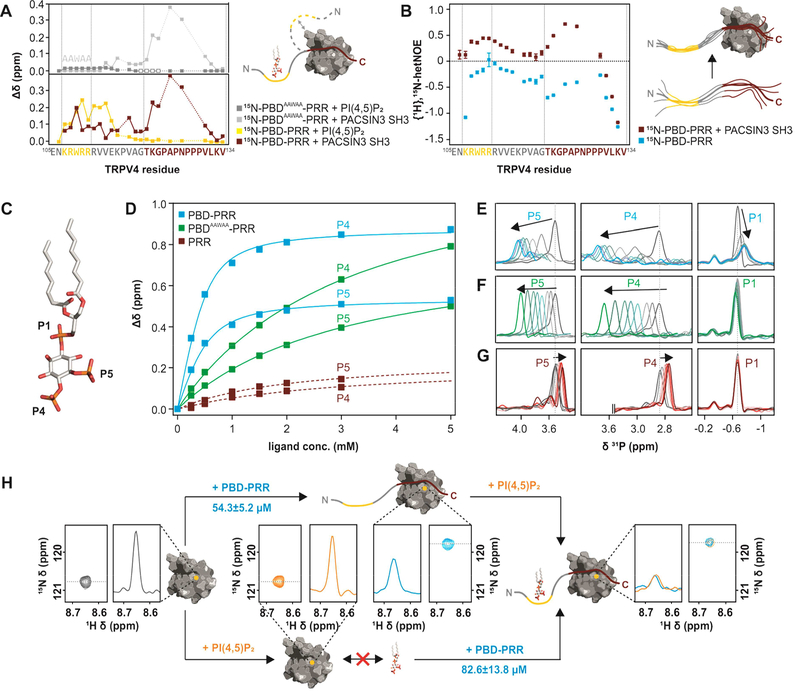
Figure 4: Interactions between TRPV4 N-terminus, PACSIN3 SH3 domain and PIP2.
(A) Chemical shift differences of TRPV4-PBD-PRR (bottom) and TRPV4-PBDAAWAA-PRR (top) free and bound states with PACSIN3 SH3 (red and light grey traces) and PIP2 (yellow and dark grey traces). PIP2 only leads to chemical shift changes in the previously determined PIP2 binding domain (Garcia-Elias et al., 2013), while PACSIN3 SH3 domain has effects on both the PRR and the PBD. For PBDAAWAA-PRR, mostly the same residues are affected by PACSIN3 SH3 binding, while PIP2 binding leads to line broadening in the linker region (empty boxes) and only very slight chemical shifts in the PBD. (B) {1H},15N-HetNOE values of TRPV4-PBD-PRR backbone amides. Lower values are indicative of higher flexibility. (C) Position of the three phosphate groups, P1, P4 and P5 in PI(4,5)P2. (D) 31P titration curves of PIP2 P4 and P5 with PBD-PRR (blue), PBDAAWAA-PRR (green) and PRR (red) derived from 31P chemical shift changes: (E-G) 31P NMR spectra of PIP2 phosphate groups with (E) PBD-PRR; (F) PBDAAWAA-PRR and (G) PRR. (H) Effect of adding PIP2 and PBD-PRR to 15N-PACSIN3 SH3 domain in different orders:Peak corresponding to PACSIN3 SH3 Y387 is shown in the 2D 1H, 15N-spectrum and as a slice through the nitrogen dimension to illustrate line width (left, grey)Upon addition of PIP2 to the SH3 domain, no chemical shift differences nor line broadening is observed, thus showing that lipid and SH3 domain do not interact (bottom, orange). In contrast, addition of PBD-PRR to 15N-PACSIN3 SH3 shows a chemical shift and concomitant line broadening indicative of interaction between SH3 domain and peptide (top, blue). In a second step, the respective missing binding partner (PIP2 in upper and PBD-PRR in lower path) is added. In both cases, identical final chemical shift positions and broad line widths are observed (right) showing that PACSIN3 SH3, PIP2 and TRPV4-PBD-PRR form the same tripartite complex regardless of order of binding partner addition.