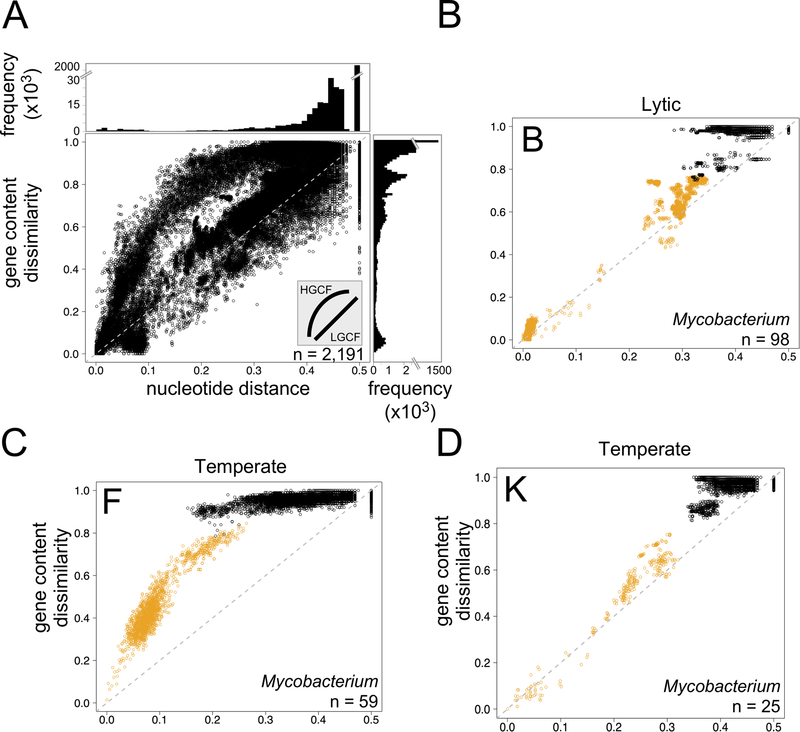
Figure 9. Modes of phage evolution.
A. Nucleotide distance (using Mash) and gene content dissimilarity (using phams from Phamerator) are plotted for ~2.4 × 106 dsDNA phage comparisons to reveal two evolutionary modes, HGCF and LGCF (inset). The line at y=2× is plotted for reference. Marginal frequency histograms emphasize densely plotted regions, with truncated y axes for viewability. B-D. Cluster-specific intra-cluster (orange) and inter-cluster (black) comparisons are plotted as in Fig. 1a for actinobacteriophage Clusters B (panel B), F (panel C), and K (panel D). Phages in Clusters F and K are temperate, and phages in Cluster B are lytic, as indicated. n, number of phages present in the specific cluster. Reproduced with permission from reference 81.