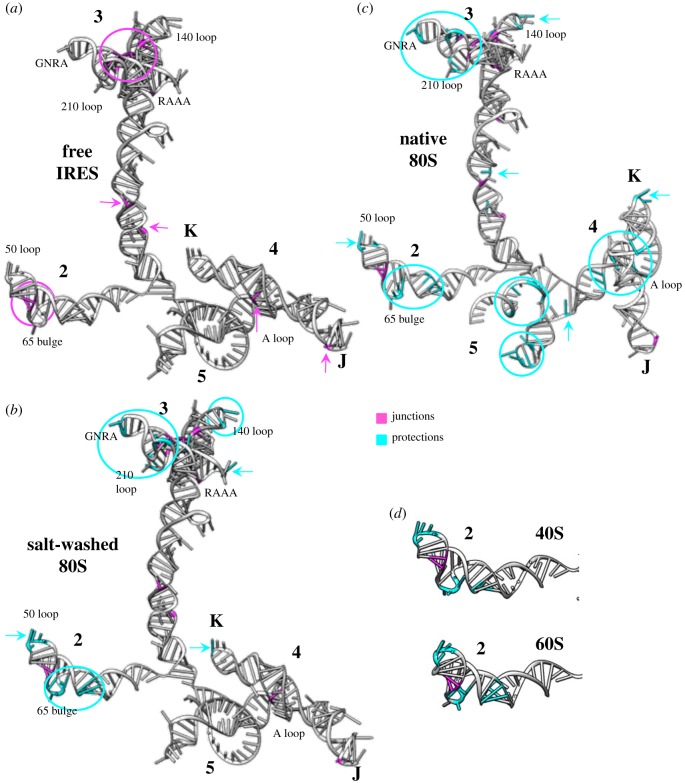
Figure 2.
Conformational changes on the FMDV IRES induced by ribosomal fractions. Three-dimensional structure models for the IRES were predicted imposing SHAPE reactivity values obtained for the free RNA. The junctions defined by statistically significant reactivity towards di-ruthenium compound footprint are highlighted in pink (a), while the statistically significant reduction of SHAPE reactivity (protections) upon incubation with the ribosomal fractions is highlighted in cyan (b). Conformational changes observed by differential SHAPE, using isatoic anhydride (IA) treatment, upon incubation of the FMDV IRES with salt-washed 80S ribosomes (b), and native 80S (c). (d) Conformational changes observed in domain 2 upon incubation with purified 40S or 60S subunits. Domains 2, 3, 4 and 5, subdomains J and K of domain 4, as well as the GNRA tetraloop, loops and bulges referred to in the text, are indicated.