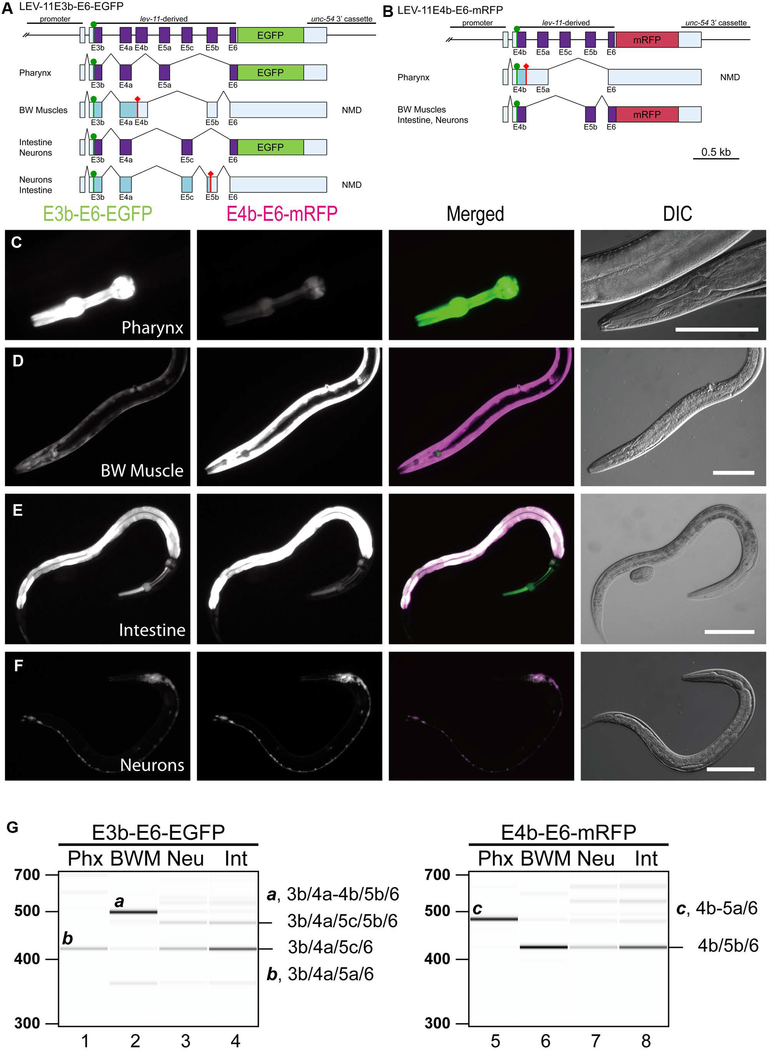
Figure 3. Fluorescence splicing reporter analysis for lev-11 E4s and E5s.
(A, B) Schematic illustration of lev-11 reporter minigenes, E3b-E6-EGFP (A) and E4b-E6-mRFP (B). Structures of major mRNA isoforms expressed in the pharynx, body wall muscles (BW Muscles), intestine, and neurons are indicated below each minigene. The cDNA cassettes and the predicted open reading frames (ORFs) for EGFP and mRFP are colored in green and magenta, respectively. Frames shown in cyan are not in-frame with the fluorescent proteins and such mRNAs are likely degraded by nonsense-mediated mRNA decay (NMD). Green circles indicate artificially-introduced initiation codons. (C-F) Fluorescence images showing expression of E3b-E6-EGFP and E4b-E6-mRFP in L4 larvae or adults under the control of the myo-2 (pharynx) (C), myo-3 (body wall muscle) (D), gst-42 (intestine) (E), or rgef-1 (neurons) (F) promoter. Micrographs were taken with the same exposure settings for the four strains. Merged images of EGFP in green and mRFP in magenta and DIC images are also shown. Bars, 100 μm. (G) RT-PCR analysis of polyadenylated mRNAs from E3b-E6-EGFP (lanes 1–4) and E4b-E6-mRFP (lanes 5–8). Major cDNA species were cloned, sequenced, and identified exon combinations are indicated (also shown schematically in A and B). These reporters were analyzed in NMD-deficient smg-2 mutant background. DNA size markers are shown on the left of lanes 1 and 5. The presented results were consistent in two independent transgenic strains.