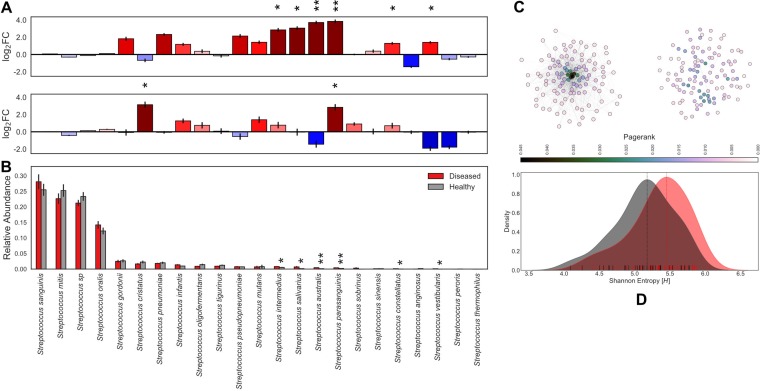
FIG 2.
Streptococcus community composition in the context of health and disease. Streptococcus community abundance analysis and enrichment in phenotype-specific cohorts. Statistical significance (P < 0.001, ***; P < 0.01, **; and P < 0.05, *). Error bars represent SEM. (A) Mean of pairwise log2 fold changes between phenotype subsets. (Top) Caries-positive versus caries-negative individuals with red indicating enrichment of taxa in the caries-positive cohort. (Bottom) Subjects with caries that has progressed to the dentin layer versus enamel-only caries with blue denoting taxa enriched in the enamel. Pseudocount of 1e−4 applied to entire-count matrix for log transformation. (B) Relative abundance of MIDAS counts for each Streptococcus species grouped by caries-positive (red) and caries-negative (gray) subjects. (C) Fully connected undirected networks for diseased (right) and healthy (left) groups separately. Edge weights represent topological overlap measures, nodes colored by PageRank centrality, and the Fruchterman-Reingold force-directed algorithm for the network layout. (D) Kernel density estimation of Shannon entropy alpha diversity distributions for caries-positive (red) and caries-negative (gray) subjects calculated from normalized Streptococcus community composition. Vertical lines indicate the mode of the kernel density estimate distributions for each cohort. Statistical significance (P = 0.008).