Figure 1.
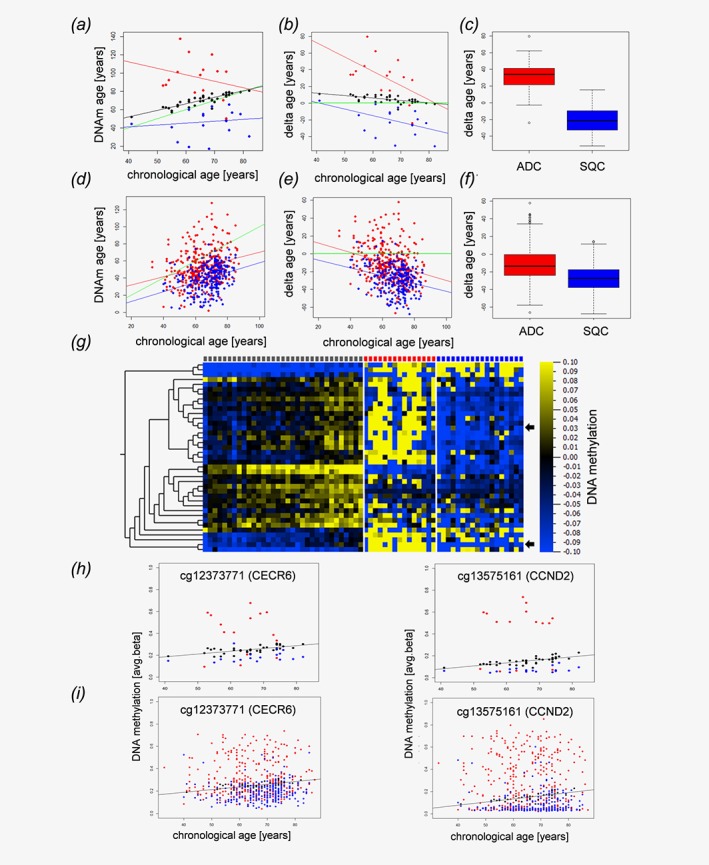
NSCLC tissues display epigenetic age distortions. Dot plot (a) and Bland–Altman‐Plot (b) showing correlations between the chronological age of the host and the epigenetic DNAm age of matched tumor tissues from normal control specimens (black dots), adenocarcinomas (red) and squamous cell carcinomas (blue). (c) Boxplot of the deviations of DNAm age from the chronological age in adenocarcinomas (ADC, red) and squamous cell carcinomas (SQC, blue). The delta age of cancer entities differs significantly (p‐value<1.32x10−5, Wilcoxon rank sum test). Validation cohort (public available TCGA data set): Dot plot (d) and Bland–Altman‐Plot (e) showing correlations between the chronological age and the epigenetic DNAm age of adenocarcinomas (red) and squamous cell carcinomas (blue) in an independent validation cohort. (f) The delta age of adenocarcinomas (ADC, red) and squamous cell carcinomas (SQC, blue) differs significantly (P‐value<2.2 x 10−16, Wilcoxon rank sum test). The delta age is calculated as DNAm age (years)—chronological age (years). (a, b, c, d) Black, red and blue lines represent the regression lines for control samples, adenocarcinomas, and squamous cell carcinomas, respectively. The green line indicates a perfect match of chronological age and DNAm age (chronological age = DNAm age). DNA methylation changes of distinct CpG loci associated with aging and alterations of these patterns in lung cancer (g–i). Heatmap (g) of CpG loci with changing DNA methylation during aging. Pearson correlation analysis of avg. beta values of all CpG loci included in our study in normal lung tissue (controls) with the chronological age of the sample donors identified 39 loci with │Pearson correlation coefficients│ > 0.6 (Table 1). The DNA methylation values of these loci are presented separately for controls (gray squares, top lane), adenocarcinoma (red squares) and squamous cell carcinoma (blue squares). Samples are ordered according to donors’ chronological ages. Heatmap: blue, low DNA methylation values; yellow, high DNA methylation values, for visualization purposes, the DNA methylation values were normalized to zero for each locus, and the bar below the heatmap indicates the range and color code. Arrows indicate randomly selected loci detailed in H and I. (h) Detailed presentation of data from two randomly selected loci. DNA methylation of cg12373771 (CECR6) and cg13575161 (CCND2) increases during aging in control samples (black spheres). Compared to the controls, squamous cell carcinoma (blue spheres) are characterized by lower DNA methylation values, while DNA methylation is increased in adenocarcinoma (red spheres), putatively reflecting decelerated and accelerated aging, respectively. (i) A similar pattern is seen also in an independent dataset.
