Figure 2.
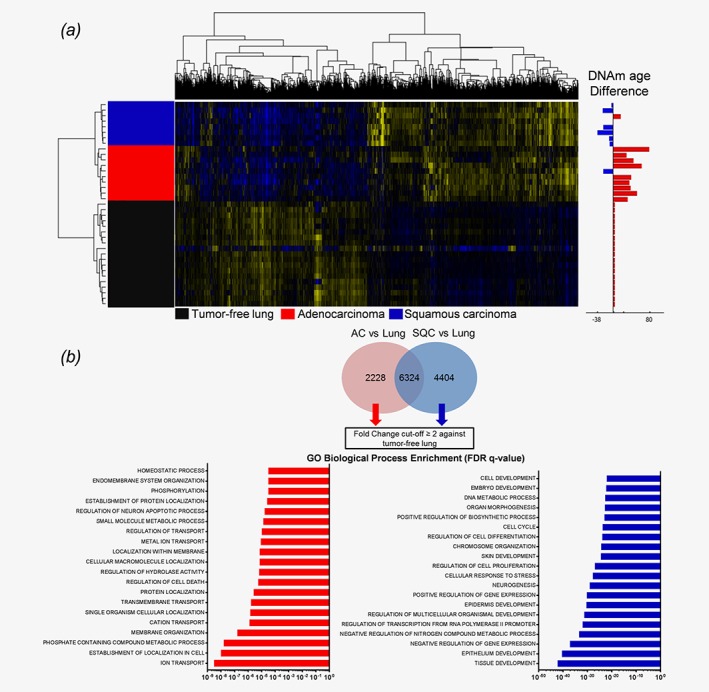
NSCLC tumors display a differential gene expression pattern associated with differentiation and developmental processes in SQC. (a) NSCLC tumors and matched tumor‐free lungs were analyzed by 1 W‐ANOVA for the significantly different expression of genes. The difference in DNAm age for each tumor compared to the chronological age of the host is displayed as an adjacent annotation track. Hierarchical clustering clearly separated all groups. (b) Specific gene expression signatures of AC and SQC tumors were derived from genes found to be significantly differentially expressed between adenocarcinomas and tumor‐free lungs and between squamous cell carcinomas and tumor‐free lungs, as indicated by the Tukey HSD post hoc test. From both gene lists, shared genes were excluded, and specific signatures were extracted. Only those genes with a fold‐change of ≥ 2 compared to the tumor‐free lung tissues were subjected to GO enrichment analysis. Gene Ontology analysis displays the top 20 most significantly enriched GO terms, as determined by the FDR q‐values.
